Abstract
T cell-mediated immune response is essential for host defense against viruses, including the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which has caused a global pandemic. Genetically engineered T cells with antigen-specific T cell receptors (TCR-Ts) are redirected to eliminate target cells via TCRs recognizing peptides bound with major histocompatibility complexes (MHCs). TCR-T cell therapy has proved effective in human trials involving solid tumors, while it is considered promising treatment for infectious diseases. To expedite the identification of functional TCRs and development of TCR-T cell therapy, we established an experimental pipeline to generate functional virus-specific TCR-Ts through coupled analysis of single-cell transcriptomic data and single-cell full length TCR V(D)J sequences. We validated this approach in selecting functional TCRs specific to a cytomegalovirus (CMV) pp65 epitope, NLV495-503, and published the results recently. In this commentary, we summarized the single-cell work flow and discussed its potential application in developing TCR-T cell therapy against SARS-CoV-2.
Keywords
TCR-T, Single-cell RNA sequencing, Single-cell TCR V(D)J sequencing, SARS-CoV-2
Introduction
Acute or chronic infectious diseases caused by human transmissible viruses (such as cytomegalovirus (CMV), severe acute respiratory syndrome coronavirus (SARSCoV), middle east respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 causing the ongoing COVID-19 pandemic) have posed tremendous threats to human health [1-4]. During viral infection, immune systems are activated to defend against viruses, including the innate immune responses, which respond rapidly as the first line of defense, and followed by the adaptive immune responses, which are composed of humoral immunity and cell-mediated immunity. In humoral immune responses, antibody-secreting effector B cells secrete neutralizing antibodies, which bind to viruses by the antigenic domain and mainly interfere with the process that virus particles entering into the host cells [5,6]. Besides, cell-mediated immune responses are also crucial to both acute and chronic viral infection. Both cytolytic activity against viral infected cells and production of immune inflammatory factors by CD4+/CD8+ T cells contribute to the clearance of viral infection. However, it was found that functional exhaustion of CD4+/CD8+ T cells help viruses to escape from T cell responses in chronic and acute viral infection [7,8]. As a consequence, effective and specific tools targeting to viruses are needed to strengthen our antivirus capabilities.
Immunotherapies, including adoptive T cell therapy, have emerged to the front line of treatment for cancers, and they are also feasible and promising as treatment options for infectious diseases [9,10]. T cell receptor engineered T cells (TCR-Ts) are T cells transferred with exogenous antigen-specific TCRs, which are redirected to recognize target cells via TCR-CD3 cluster binding to the peptide- MHC. After recognition, downstream signaling pathways are activated to release cytokines and granzymes (such as IFN-γ, TNF-α, granzyme A and granzyme B) to execute cytotoxicity to target cells and activate other immune cells [11]. In essence, TCR-T cell therapy harnesses our immune system to control viral duplication and attack infected cells or tumors in patients.
Single-cell Approach to Generate Functional TCR-Ts
A major technical problem in the development of TCR-T cell therapy comes from accurate identification of functional TCRs with paired α and β chains [12]. Advances of next-generation sequencing technology enables highthroughput parallel sequencing of millions of DNA fragments, however, sequencing from bulk materials usually lost the information on the pairing of TCR α and β chains. Single-cell full-length TCR sequencing, along with single-cell RNA sequencing opens up a new path to map the TCR repertoire with paired α and β sequences [13,14], which can be achieved through several technologies and platforms, such as microdroplet-based microfluidics technology [15], Smart-seq2 technology [16], nested PCR [17], VDJ Puzzle [18], and Single-cell TCR seq [19]. Besides the TCR repertoire profiling, the correlated analysis of the single-cell transcriptome and TCR information allows for functional TCR selection based on transcription profiles, which have not been fully explored yet.
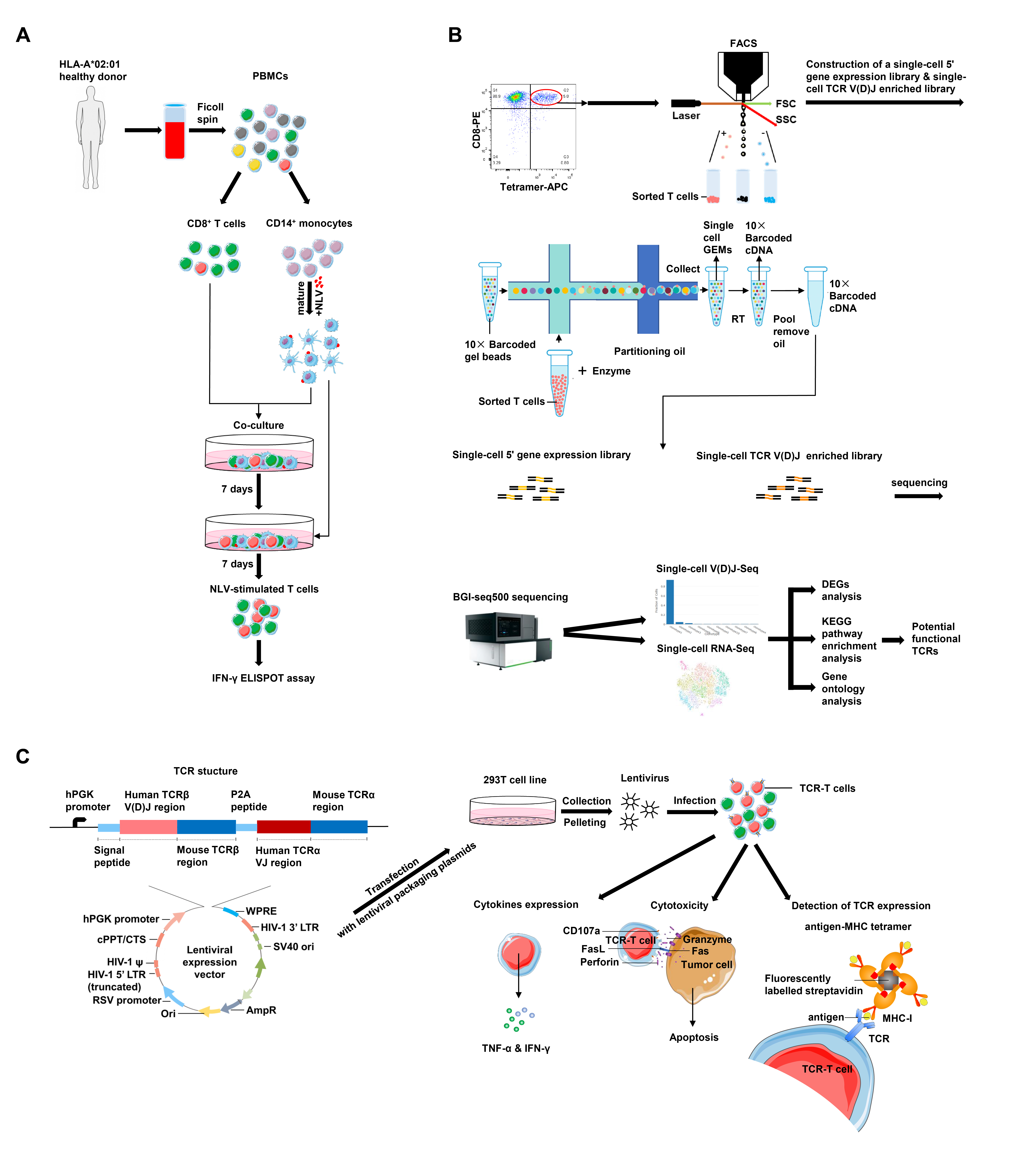
Our recent work performed the single-cell transcriptome and single-cell TCR analysis in selecting functional TCRs specific to CMV immunodominant epitope NLV495-503 and established a comprehensive pipeline to generate functional TCR-Ts [20]. Most individuals infected by CMV usually remain asymptomatic due to their robust immune response [21]. However, CMV reactivation occurs frequently in immunocompromised patients or immunocompetent patients with critical illness [22,23]. Reconstruction of a functional CMV-specific T cell repertoire has been shown to effectively control CMV-related diseases [9,24,25]. In our research, NLV495-503-specific T cells were preferentially expanded in vitro by co-culture and stimulation of primary CD8+ T cells from a healthy donor with NLV495- 503 peptide-loaded autologous dendric cells (DCs) (Figure 1A). The specific reactivity to the antigen was measured by the IFN-γ enzyme-linked immunospot (ELISPOT) assay. NLV495-503-specific CD8+ T cells were then stained by the fluorescently labeled NLV495-503-MHC tetramer and sorted (Figure 1B). Single cells were isolated by microdropletbased microfluidics, followed by construction of the singlecell 5’ gene expression library and the single-cell TCR V(D)J enriched library, and sequencing on a BGI-seq 500 platform. Single-cell RNA sequencing of T cells allowed grouping of cells with similar transcriptomic features into clusters by unsupervised clustering. Meanwhile, paired, full-length TCR sequences were obtained from singlecell TCR sequencing and TCR clonotypes were mapped onto the clusters according to the transcriptomic data. As a result, 2 clusters with distinct transcriptomic features were defined, and further bioinformatic analysis suggested that cluster 1 were potentially functional T cells with high expression of cytotoxic effector molecules (GZMH and NKG7), proinflammatory cytokine IFNG, exhaustion marker TIGIT and 40 highly-expressed functional genes involved in cytokine receptor–ligand interactions as well as lymphocyte activation, while cluster 2 had features of naïve populations with high expression of naïve cell-related markers (CD27 and LEF1). Representative TCR clonotypes were selected from both clusters and corresponding TCR-Ts were generated from primary T cells infected with lentiviral vectors carrying TCR expression cassettes (Figure 1C). In vitro assays were carried out to verify TCR expression, cytokine production and cytotoxic activity of TCR-Ts. In accordance with bioinformatic predictions, the TCR from cluster 1 was highly effective in functional assays and preliminarily potential for TCR-T cell therapy, while the TCR from cluster 2 was nonfunctional. Thus, we provided a single-cell approach to select functional TCRs, which is promising to be used in effective screening of antigen-specific TCRs and valid construction of functional TCR-Ts.
T cell Immune Response to SARS-CoV-2
The pandemic of COVID-19, caused by a novel coronavirus, SARS-CoV-2, has led to a human health crisis globally. As of July 22, 2020, there have been more than 14 million people infected, including over 612 thousand deaths reported to WHO throughout the world (data from WHO, https://covid19.who.int/). Understanding the host immune response and immunopathology of COVID-19 is critical for the proper use of clinical care, interpretation of clinical outcomes, development of effective therapies, and calibration of prevention measures. With rapidly emerging research work on this matter, we are now getting more profound knowledge of the innate and adaptive immune responses involved in COVID-19 [26].
A growing body of evidence suggested that apart from the humoral immune response, T cell-mediated immune response plays essential roles in the defense against SARS-CoV-2. A study of non-severe COVID-19 reported that similar to antibody secreting cells, the frequency of activated CD38+HLA-DR+ T cells in the patient’s blood increased rapidly before the symptoms resolved, and remained higher than healthy controls days during convalescence, which demonstrated the induction of adaptive immune response by viral infection [27]. Characterization of bronchoalveolar lavage fluid (BALF) immune cells from COVID-19 patients by single-cell RNA sequencing revealed that a larger proportion of CD8+ T cells were present in BALFs from patients with moderate disease compared to patients with severe COVID-19 [28]. Furthermore, CD8+ T cells in moderate cases contained much more highly expanded TCR clones, indicating that they might function in controlling the disease progression. A longitudinal study also observed expanded or contracted T cell clones in PBMCs from two patients with mild COVID-19 and suggested that these clones were associated with the infection, most of which developed memory phenotypes by day 30 after the symptom onset [29].
T cells reactive to the components of SARS-CoV-2, especially the spike protein epitopes have been identified in PBMCs from COVID-19 patients [30-32]. Peptide megapools were generated for stimulation of CD8+ or CD4+ T cells from the patients, corresponding to HLA class I and HLA class II epitopes predicted by bioinformatic tools covering the entire viral proteome [31]. T cell activation markers, including CD137, CD69, and OX40, were analyzed by flow cytometry to identify specific T-cell response after stimulation. 100% of the convalescent COVID-19 patients displayed CD4+ T cells reactive to the peptide megapools while the majority of them had specific response in CD8+ T cells, albeit the frequency of reactive T cells was low, generally below 1% of the CD4+ or CD8+ population. The frequency of spike-reactive CD4+ T cells was corelated with the spike-specific antibody titer in COVID-19 patients, in line with the functional role of CD4+ T cells in promoting B cell maturation and antibody production [33]. Surprisingly, CD4+ and CD8+ T cells from a portion of healthy donors showed cross-reactivity to the predicted SARS-CoV-2 epitopes, which might result from their previous contact with other human coronaviruses [30,31]. Whether this pre-existing cross-reactive T cells helped in protecting from SARS-CoV-2 infection remains to be validated in a larger number of subjects.
Potential of TCR-T cell Therapy against SARS-CoV-2
Great efforts have been made to develop therapeutics and vaccines against SARS-CoV-2. For example, neutralizing antibodies targeting the spike protein on the viral surface have shown strong efficacies in prevention of viral infection in vitro and in mouse models, making them promising drug candidates [34,35]. As for vaccine approaches, studies showed that vaccines induced protective immunity including potent neutralizing antibodies and CD8+ T cell responses, providing efficient protection against SARSCoV- 2 in mice and rhesus macaques [36,37]. Clinical trials are being held worldwide to evaluate the safety and efficacy of the drug candidates and vaccines.
A study showed that scientists identified memory T cell clones reactive to SARS-CoV-1, a prior coronavirus closely related to SARS-CoV-2, from individuals at 6 years post infection, and generated TCR-Ts transduced with the most potent TCR, which preserved the functions of the parent T cell clone and responded to viral antigen stimulation in vitro [38]. Though functional TCR-Ts have not yet been reported for SARS-CoV-2, TCR-T cell therapy holds the potential to become a new prophylactic or therapeutic treatment option to fight the current coronavirus outbreak [39].
There are two main strategies to identify TCRs for SARS-CoV-2 and assess their functions. First, one could monitor the dynamics of T cell clones through longitudinal TCR repertoire profiling and identify TCR clones that are expanded or contracted after SARS-CoV-2 infection [29]. For hundreds of TCR clones significantly expanded or contracted, computational analysis was performed to cluster TCR sequences, with each cluster containing highly similar clonotypes. Characteristic TCR motifs were identified, some of which were shared between different donors with COVID-19, suggesting that there might be public TCRs of diagnostic or therapeutic value. Yet the analysis is basically in silico, generation of TCR-Ts and functional assessment is necessary to determine whether these TCRs are truly reactive to SARS-CoV-2.
The second approach is to identify T cell clones reactive to SARS-CoV-2 by stimulation with viral immunogenic peptides [31,32]. It could be conveniently adapted to our functional TCR-T generation pipeline (Figure 1). Since virus-reactive T cell clones are also present in healthy donors [30,31], it should be possible to make use of CD8+ T cells sorted from patient-derived PBMC or from healthy donors. In-vitro priming of T cells with autologous DCs loaded with immunogenic viral peptides allows for efficient expansion of virus-specific T cell clones [40], which would be detected by the corresponding tetramer staining and isolated for single-cell TCR sequencing and RNA sequencing. The concurrent analysis of single-cell transcriptome and TCR clonotypes would provide more thorough information on selection of functional TCRs than a few conventional activation markers [30-32], and thereby streamline the downstream experiments including generation of TCR-Ts and functional assessment.
Limitations of TCR-T cell Therapy in Infectious Diseases
Several major challenges remain for the applications of TCR-T cell therapy in infectious diseases. Firstly, TCR recognition of the peptide-MHC is HLA-restricted, therefore one TCR-T cell therapy could not fit all, but has to target the HLA-matched patients. Secondly, safety concerns of TCR-T cell therapy need to be taken considerations, including excessive lyse of the infected cells which might be part of the essential organs and cytokine storms which might be life-threatening [41]. Lastly, TCR-T cell therapy, like other gene-modified cell therapies, requires specialized facilities and complicated production processes, which might be a big hurdle for its widely applications [42].
Nonetheless, the emerging new techniques such as highthroughput functional TCR screening [20,43], TCR-Ts with safety switches and universal TCR-Ts [44] as well as evolving manufactural technologies [45] will ultimately enhance our abilities to design and produce better TCR-T products for clinical use.
Conclusion
T cells are natural essential effector cells against virus infection. Adoptive cell therapy using virus-specific TCR-Ts has clinical applications in treating CMV-related diseases, and holds the potential to become a new prophylactic or therapeutic option for acute infectious diseases such as COVID-19. However, TCR-T cell therapy is being held back by various obstacles including lack of efficient methods for identifying functional TCRs. In our pilot study, we devised a simple and rapid approach to generate functional TCR-Ts based on the correlative analysis of the transcriptome and TCR clonotypes from single-cell RNA and full-length TCR sequencing data. Further investigations are in progress to validate its applications in functional TCR-T generation for other immunogens.
Acknowledgement
This project is supported by the Science, Technology and Innovation Commission of Shenzhen Municipality under grant No. JCYJ20170303151334808 and grant No. JSGG20180508152912700.
Declaration of Interests
The authors declare no competing interests.
References
2. Peiris JSM, Yuen KY, Osterhaus ADME, Stöhr K. The Severe Acute Respiratory Syndrome. New England Journal of Medicine. 2003;349(25):2431-41.
3. Arabi YM, Balkhy HH, Hayden FG, Bouchama A, Luke T, Baillie JK, et al. Middle East Respiratory Syndrome. New England Journal of Medicine. 2017;376(6):584-94.
4. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England Journal of Medicine. 2020;382(8):727-33.
5. Murin CD, Wilson IA, Ward AB. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nature Microbiology. 2019;4(5):734-47.
6. Subbarao K, Mahanty S. Respiratory Virus Infections: Understanding COVID-19. Immunity. 2020;52(6):905-9.
7. Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cellular & Molecular Immunology. 2020;17(5):533-5.
8. Kahan SM, Wherry EJ, Zajac AJ. T cell exhaustion during persistent viral infections. Virology. 2015;479- 480:180-93.
9. Feuchtinger T, Opherk K, Bethge WA, Topp MS, Schuster FR, Weissinger EM, et al. Adoptive transfer of pp65-specific T cells for the treatment of chemorefractory cytomegalovirus disease or reactivation after haploidentical and matched unrelated stem cell transplantation. Blood. 2010;116(20):4360-7.
10. Kaeuferle T, Krauss R, Blaeschke F, Willier S, Feuchtinger T. Strategies of adoptive T -cell transfer to treat refractory viral infections post allogeneic stem cell transplantation. Journal of Hematology & Oncology. 2019;12(1):13.
11. Ping Y, Liu C, Zhang Y. T-cell receptor-engineered T cells for cancer treatment: current status and future directions. Protein Cell. 2018;9(3):254-66.
12. Rosati E, Dowds CM, Liaskou E, Henriksen EKK, Karlsen TH, Franke A. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnology. 2017;17(1):61.
13. Zemmour D, Zilionis R, Kiner E, Klein AM, Mathis D, Benoist C. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nature Immunology. 2018;19(3):291-301.
14. Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. 2018;174(5):1293-308 e36.
15. Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Experimental & Molecular Medicine. 2018;50(8):96.
16. Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive fulllength transcriptome profiling in single cells. Nature Methods. 2013;10(11):1096-8.
17. De Simone M, Rossetti G, Pagani M. Single Cell T Cell Receptor Sequencing: Techniques and Future Challenges. Frontiers in Immunology. 2018;9:1638.
18. Eltahla AA, Rizzetto S, Pirozyan MR, Betz-Stablein BD, Venturi V, Kedzierska K, et al. Linking the T cell receptor to the single cell transcriptome in antigen-specific human T cells. Immunology and Cell Biology. 2016;94(6):604-11.
19. Redmond D, Poran A, Elemento O. Single-cell TCRseq: paired recovery of entire T-cell alpha and beta chain transcripts in T-cell receptors from single-cell RNAseq. Genome Medicine. 2016;8(1):80.
20. Wang F, Xu Q, Zhuang Z, Li Z, Gao Q, Huang Y, et al. A single-cell approach to engineer CD8+ T cells targeting cytomegalovirus. Cellular & Molecular Immunology. 2020;doi: 10.1038/s41423-020-0466-z. Online ahead of print.
21. Zanghellini F, Boppana SB, Emery VC, Griffiths PD, Pass RF. Asymptomatic Primary Cytomegalovirus Infection: Virologic and Immunologic Features. The Journal of Infectious Diseases. 1999;180(3):702-7.
22. Hodson EM, Jones CA, Webster AC, Strippoli GFM, Barclay PG, Kable K, et al. Antiviral medications to prevent cytomegalovirus disease and early death in recipients of solid-organ transplants: a systematic review of randomised controlled trials. The Lancet. 2005;365(9477):2105-15.
23. Limaye AP, Kirby KA, Rubenfeld GD, Leisenring WM, Bulger EM, Neff MJ, et al. Cytomegalovirus Reactivation in Critically Ill Immunocompetent Patients. JAMA. 2008;300(4):413-22.
24. Riddell, Watanabe KS, Goodrich JM, Li CR, Agha ME, Greenberg PD. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257(5067):238.
25. Schub A, Schuster I, Hammerschmidt W, Moosmann A. CMV-specific TCR-transgenic T cells for immunotherapy. Journal of Immunology (Baltimore, Md : 1950). 2009;183:6819-30.
26. Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of COVID-19: Current State of the Science. Immunity. 2020;52(6):910-41.
27. Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Caly L, van de Sandt CE, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of nonsevere COVID-19. Nature Medicine. 2020;26(4):453-5.
28. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Singlecell landscape of bronchoalveolar immune cells in patients with COVID-19. Nature Medicine. 2020;26(6):842-4.
29. Minervina AA, Komech EA, Titov A, Koraichi MB, Rosati E, Mamedov IZ, et al. 2020. Longitudinal highthroughput TCR repertoire proling reveals the dynamics of T cell memory formation after mild COVID-19 infection. bioRxiv doi: 10.1101/2020.05.18.100545
30. Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, et al. 2020. Presence of SARS-CoV-2-reactive T cells in COVID-19 patients and healthy donors. medRxiv doi: 10.1101/2020.04.17.20061440
31. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T Cell Responses to SARSCoV- 2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181(7):1489-501 e15.
32. Shomuradova AS, Vagida MS, Sheetikov SA, Zornikova KV, Kiryukhin D, Titov A, et al. 2020. SARSCoV- 2 epitopes are recognized by a public and diverse repertoire of human T-cell receptors. medRxiv doi: 10.1101/2020.05.20.20107813
33. Sette A, Moutaftsi M, Moyron-Quiroz J, McCausland MM, Davies DH, Johnston RJ, et al. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008;28(6):847-58.
34. Wang C, Li W, Drabek D, Okba NMA, van Haperen R, Osterhaus ADME, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nature Communications. 2020;11(1):2251.
35. Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182(1):73-84.e16.
36. Corbett KS, Edwards D, Leist SR, Abiona OM, Boyoglu- Barnum S, Gillespie RA, et al. 2020. SARS-CoV-2 mRNA Vaccine Development Enabled by Prototype Pathogen Preparedness. bioRxiv doi: 10.1101/2020.06.11.145920
37. Wang H, Zhang Y, Huang B, Deng W, Quan Y, Wang W, et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell. 2020;182(3):713–21.e9.
38. Oh HL, Chia A, Chang CX, Leong HN, Ling KL, Grotenbreg GM, et al. Engineering T cells specific for a dominant severe acute respiratory syndrome coronavirus CD8 T cell epitope. Journal of Virology. 2011;85(20):10464- 71.
39. Gutierrez L, Beckford J, Alachkar H. Deciphering the TCR Repertoire to Solve the COVID-19 Mystery. Trends in Pharmacological Sciences. 2020;41(8): 518–30.
40. Wolfl M, Greenberg PD. Antigen-specific activation and cytokine-facilitated expansion of naive, human CD8+ T cells. Nature Protocols. 2014;9(4):950-66.
41. Tan AT, Bertoletti A. Challenges of CAR- and TCR-T cell–based therapy for chronic infections. Journal of Experimental Medicine. 2020;217(5):e20191663.
42. Wang X, Riviere I. Manufacture of tumor- and virusspecific T lymphocytes for adoptive cell therapies. Cancer Gene Therapy. 2015;22(2):85-94.
43. Segaliny AI, Li G, Kong L, Ren C, Chen X, Wang JK, et al. Functional TCR T cell screening using single-cell droplet microfluidics. Lab on a chip. 2018;18(24):3733-49.
44. June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science. 2018;359(6382):1361-5.
45. Jin J, Gkitsas N, Fellowes VS, Ren J, Feldman SA, Hinrichs CS, et al. Enhanced clinical-scale manufacturing of TCR transduced T-cells using closed culture system modules. Journal of Translational Medicine. 2018;16(1):13.