Abstract
The enteric nervous system is the largest component of the autonomic nervous system. It contains a broad network of interconnected plexuses and enteric neuronal subtypes which are in charge of the normal functioning of the gastrointestinal tract. Vagal and sacral neural crest cells are at the basis of the enteric nervous system development. These cells undergo multiple processes such as migration, proliferation and differentiation to finally form a functional enteric nervous system. These processes are the result of multiple transcription factors, signaling molecules and pathways and their interactions, which all play essential roles in the enteric nervous system development. Malfunctioning or failing of one of these processes leads to congenital enteric neuropathies such as Hirschsprung’s disease, characterized by partial aganglionosis of the gastrointestinal tract. Since the current therapy brings many post-operative complications and a reduced quality of life, there is a high demand for alternative therapies such as stem cell therapy. To apply stem cell therapy, it is necessary that the factors involved in the differentiation process are known and thus that the process is fully understood. While migration and proliferation processes are largely unraveled, the differentiation process and its factors still remain largely unknown. This review describes the factors which have been identified so far and are involved in the differentiation and subtype specification of enteric neurons.
Keywords
Enteric nervous system, Developmental biology, Transcription factors, Signaling molecules, Signaling pathways, Enteric neuron differentiation, Enteric neuron subtype specification
Introduction
The enteric nervous system
The enteric nervous system (ENS) forms the largest component of the autonomic nervous system (ANS). In humans, it contains between 200 and 600 million neurons which are part of intrinsic neuronal circuits managing to generate reflex gastrointestinal (GI) contractile activity without intervention of the central nervous system (CNS) [1,2]. The ENS is located along the length of the GI tract and oversees controlling the main functions such as secretion, motility, and blood flow. In addition, it is also responsible for the communication with the immune system and microbiome [3–5]. The ENS contains a network of neurons and glial cells which are dispersed over two major ganglionated and interconnected plexuses, the myenteric (Auerbach) plexus, and the submucosal (Meissner) plexus. In larger mammals, the submucosal plexus is further subdivided into smaller plexuses [4,6-10]. The neurons of the myenteric plexus are primarily involved in GI motility regulation, while the neurons of the submucosal plexus are involved in the regulation of secretion and vascular tone [3,6,11,12]. The ENS is a highly complex nervous system of which the functioning is dependent on many different neuronal subtypes. To keep an overview of the neuronal subtypes, they are categorized in different classes according to certain characteristics. Among these features are their morphology, electrical properties, chemical coding, and functioning [13]. In the beginning, the enteric neurons were identified by Dogiel based on their morphology (reviewed by Brehmer) [14]. Later, other characteristics such as the ones listed above, were combined, and included in the classification. The standard nowadays is based on the neurochemical coding combined with functional experimental results, and consists of the following groups: intrinsic primary afferent neurons (IPANS, sensory neurons), motor neurons, interneurons, intestinofugal neurons, and secretomotor and vasomotor neurons [3,6,13].
Throughout the years, research is performed on the identification of different neuronal subtypes using multiple experimental animal models. The animal models share in some part the genetic coding of the enteric neuronal subtypes. Nevertheless, we must keep in mind that even between closely related species, differences in neurochemical coding and morphology occur [6]. The most popular models are the guinea pig, mouse, and zebrafish. In the small intestine of the guinea pig, research on neuronal diversity resulted in the identification of 14 functionally important subtypes. This is reviewed by Furness and Hao & Young (Table 1) [7,11]. These subtypes belong to one of the classes listed above, which are equally divided along the entire network of ganglia [3,7,11]. In the zebrafish, at least 10 neuronal subtypes are defined based on their neurochemical coding. The combination of different neuronal markers and comparisons with previous described enteric neuronal functions in other animal models led to appointing putative functions to these different subtypes (Table 2) [15,16]. Recently, a novel taxonomy of 12 myenteric neuron classes of the mouse small intestine was introduced (Table 3). These neurons are defined by their communicating characteristics portrayed along with neurochemical markers and transgenic tools. After defining the genetic codes of the neuronal classes, again comparisons were performed with previous studies to possibly identify the functions of each class [5]. The selective expression of genes conferring neuronal properties strongly suggests that these 12 classes represent functionally distinct neurons [5]. In the human ENS, subpopulations of enteric neurons are also characterized by their differential expressed neurotransmitters, neurotransmitter synthesis enzymes, and other molecules [11]. Initially, at least 9 myenteric major neuron classes were identified based on their morphological and neurochemical characteristics [14]. Recently, the number of enteric neuronal subtypes increased to 22 throughout the entire human GI tract. In addition, it is shown that these subtypes represent distinct, regional expression patterns of the marker genes, which opens the road towards regional specific therapeutics [17].
| Neuronal class | Chemical coding | Neuronal function |
|---|---|---|
| 1 | ChAT/TK/ENK/GABA ChAT/TK/ENK/NFP |
Excitatory circular muscle motor neuron |
| 2 | NOS/VIP/PACAP/ENK/NPY/GABA NOS/VIP/PACAP/DY/BN/NFP |
Inhibitory circular muscle motor neuron |
| 3 | ChAT/Calr/TK | Excitatory longitudinal muscle motor neuron |
| 4 | NOS/VIP/GABA | Inhibitory longitudinal muscle motor neuron |
| 5 | ChAT/Calr/TK | Inhibitory longitudinal muscle motor neuron |
| 6 | ChAT/NOS/VIP ± BN ± NPY | Descending interneuron (local reflex) |
| 7 | ChAT/5-HT | Descending interneuron (secretomotor reflex) |
| 8 | ChAT/SOM | Descending interneuron (migrating myoelectric complex) |
| 9 | ChAT/Calb/TK/NK3 receptor | IPAN |
| 10 | ChAT/BN/VIP/CCK/ENK | Intestinofugal neuron |
| 11 | VIP/GAL | Non-cholinergic secretomotor/vasodilator neuron |
| 12 | ChAT/Calr/DY | Cholinergic secretomotor/vasodilator neuron |
| 13 | ChAT/NPY/CCK/SOM/CGRP/DY | Cholinergic secretomotor/non-vasodilator neuron |
| 14 | ChAT/TK/Calb | IPAN |
Abbreviations: ChAT: Choline Acetyltransferase; TK: Tachykinin; ENK: Enkephalin; GABA: Gamma Amino Butyric Acid; NFP: Neurofilament Protein; NOS: Nitric Oxide Synthase; VIP: Vasoactive Intestinal Peptide; PACAP: Pituitary Adenylyl Cyclase Activating Peptide; NPY: Neuropeptide Y; DY: Dynorphin; BN:Bombesin; Calr: Calretinin; Calb: Calbindin; 5-HT: 5-Hydroxy Tryptamine or Serotonin; GAL: Galanin; CCK: Cholecystokinin; SOM: Somatostatin
Table 1: The different neuronal classes with their chemical coding and function in the gastrointestinal tract of the guinea-pig [7].
| Neuronal class | Neurochemical coding | Putative function |
|---|---|---|
| 1 | 5-HT | Motor or secretomotor neurons |
| 2 | 5-HT/VIP/PACAP | Vasodilator or secretomotor neurons |
| 3 | Calb/Calr/ChAT/nNOS | Motor or sensory neurons |
| 4 | Calb/Calr/nNOS | Sensory or interneurons |
| 5 | Calb/Calr/nNOS/VIP/PACAP | Inhibitory motor neurons |
| 6 | Calb/Calr/ChAT | Excitatory motor or sensory neurons |
| 7 | VIP/PACAP | Vasodilator or secretomotor neurons |
| 8 | ChAT | - |
| 9 | ChAT/GAL | Interneurons |
| 10 | GAL | Vasodilator or secretomotor neurons |
Abbreviations: 5-HT: 5-Hydroxy Tryptamine or Serotonin; VIP: Vasoactive Intestinal Peptide; PACAP: Pituitary Adenylyl Cyclase Activating Peptide; Calb: Calbindin; Calr: Calretinin; ChAT: Choline Acetyltransferase; nNOS: Neuronal Nitric Oxide Synthase; GAL: Galanin.
Table 2: Summary of the enteric neuronal subtypes identified in zebrafish and their putative functions [15,16].
| Neuronal class | Genetic coding | Neuronal function |
|---|---|---|
| 1 | Tac1, Cxcl12, Calb2, Ndufa4l2 | Excitatory motor neurons |
| 2 | Tac1, Calb2, Ndufa4l2, Gda, Penk | Excitatory motor neurons |
| 3 | Tac1, Ndufa4l2, Gda, Penk, Nxph1 | Excitatory motor neurons |
| 4 | Tac1, Gda, Penk, Fut9, Nfatc1 | Excitatory motor neurons |
| 5 | Som | Interneuron 2 |
| 6 | Nmu | IPANs |
| 7 | Cck, Ucn3 | IPANs |
| 8 | Nos1,C1q11,Npy, Cox8b | Inhibitory motor neurons |
| 9 | Nos1, C1q11, (Npy (<<8)), (Cox8b (<<8)) | Inhibitory motor neurons |
| 10 | NeuroD6, Nos1 | Interneuron 1 |
| 11 | Npy,Dlk1 | - |
| 12 | Nxph2 | IPANs Subset: Interneuron 3 |
Abbreviations: Tac1: Tachykinin Precursor 1; Cxcl12: C-X-C motif chemokine ligand 12; Calb2: Calbindin 2; Ndufa4l2: NADH dehydrogenase 1-alpha subcomplex 4-like 2; Gda: Guanine deaminase; Penk: Proenkephalin; Nxph1: Neurexophilin 1; Fut9: Fucosyltransferase 9; Nfatc1: Nuclear factor of activated T-cells 1; Som: Somatostatin; Nmu: Neuromedin u; Cck: Cholecystokinin; Ucn3: Urocortin 3; Nos1: Nitric oxide synthase 1; Npy: Neuropeptide Y; Cox8b: Cytochrome c oxidase subunit 8b; NeuroD6: Neurogenic differentiation factor 6; Dlk1: Delta like non-canonical notch ligand 1; Nxph2: Neurexophilin 2
Table 3: Summary of the novel classification of myenteric neuron classes of the mouse small intestine with their genetic coding and neuronal function [5].
ENS development and enteric neuron differentiation
During embryogenesis, the ENS originates from vagal and sacral neural crest cells (NCCs). While the vagal NCCs are colonizing the entire length of the GI tract, the sacral NCCs show a small contribution to the ENS by colonizing the postumbilical region [9,18]. Between embryonic day (E) 8.5 and 9.5 in mice, and before the 4th week of pregnancy in humans, NCCs delaminate from the neural crest and become progenitors for multiple cell types among which the enteric neurons [11,19-21]. After delamination, the vagal NCCs start to migrate towards the developing foregut. Once arrived, they are called enteric neural crest cells (ENCCs) and are characterized by the expression of multiple factors, such as Sox10, Phoxb2, Ednrb and Ascl1. The ENCCs will then further migrate in the GI tissue, proliferate, and colonize the entire length of the GI tract and form neuronal and glial precursors [9,21,22]. The sacral NCCs migrate ventrally and will form extrinsic pelvic ganglia. From there, they will migrate into the GI tract where they will contribute to the production of enteric neuronal and glial precursors [9]. By the 7th week of human embryogenesis, the ENCCs reach the hindgut [11,22]. It is at this point that in the mouse and chicken models, the sacral NCCs enter the hindgut as well. However, the sacral NCCs are shown to colonize the hindgut independently from the vagal NCCs [23]. In a recent study, it is seen that there is a subset of the vagal NCCs that migrates through the GI mesentery to complement the ENS formation. This information has led to the introduction of a new model in ENS development. First, the vagal NCCs migrate into the dorsal mesentery of the foregut. Next, most of these cells, which are in this model called the ENCCs, enter the foregut and migrate further along the GI tract. The small remaining portion of the vagal NCCs, the mesenteric NCCs (MNCCs), will continue to migrate along the mesentery. During their migration, the MNCCs invade the GI tract to provide enough ENCCs for ENS formation. The MNCCs show a relatively slow migration, due to which the regions they invade are already occupied by the ENCCs. This makes ENCCs and MNCCs indistinguishable from each other [24].
During and after migration, the differentiation process is initiated during which ENCCs are committed to different neuronal fates [11,22]. This process is asynchronous since the appearance of neuronal subtype markers is staggered, and continues into postnatal stages [11,25]. During neuron differentiation, a reduction of Sox10-expression is observed, while the expressions of Ednrb, Ascl1 and Phoxb2 are maintained. With the decrease of Sox10-expression, comes the upregulated expression of the pan-neuronal markers HuC/D, β-tubulin III (also referred to as TuJ1) and neurofilament M [11,22]. Of these pan-neuronal markers, the protein HuC/D is routinely used in both the CNS and the peripheral nervous system (PNS). The advantages of this marker is that it is not only expressed in all neurons and thus not only in certain subpopulations. It also does not label neuronal processes, making quantification of neurons easier [26]. The development of the different neuronal subtypes occurs in a temporally sequential matter. This specific order suggests that the differentiation capacities of ENCCs is changing over time. This change is most likely to be determined by the presence of multiple factors which are largely undetermined so far [25].
With the formation of the ENS, it sometimes happens that one of the essential processes (migration, proliferation, differentiation) is failing. These processes are defined by genetically programmed pathways in which genes are expressed in precise spatiotemporal patterns regulated by gene regulatory networks. These networks are modular feedforward and feedback mechanisms comprising some subcircuit classes. They provide system-level views of the organogenesis, but are also at the genetic basis of developmental diseases through mutations [27–29]. This leads to a number of congenital enteric neuropathies, such as Hirschsprung’s disease (HSCR) [21,22,30]. HSCR is an enteric neuropathy which occurs in 1:5000 live-births and is characterized by aganglionosis along variable lengths of the distal intestine [10,20]. In most cases (80-90%), aganglionosis is affecting the rectum and the distal part of the sigmoid colon [20,21,32]. HSCR is caused by the failing of ENCCs to colonize the distal part of the intestinal tract during gestation (weeks 4 to 12 of human embryonic development). The genes undergoing mutations playing a role in the development of HSCR, predominantly belong to one of the following signaling pathways: Ret/Gdnf/Ntn and Ednrb/Et-3/Ece-1 [28]. The genetic susceptibility from multiple genes involved in this disease is widespread and variable, which reflects in differing presentation and recurrence risks among relatives [31]. A rare HSCR-variant, the so-called skip segment HSCR, contains an area within the aganglionated GI tissue in which the intestine shows the presence of ganglia, therefore containing enteric neurons and glial cells [24]. HSCR leads to functional obstruction and an impaired GI motility which is potentially fatal [10,32]. So far, the therapy applied consists of a surgical resection of the aganglionated GI tissue and anastomosis of healthy innervated tissue with the remaining part of the rectum [21,32,33]. The functional outcome of the treatment is variable, and many post-operative complications are involved. This leads to a reduced quality of life of the patients [32]. Alternative therapies such as stem cell therapy are therefore desired in the treatment of HSCR and other congenital enteric neuropathies.
An increased focus on the development and evaluation of stem cell-based therapies has been arising over the last decade in search of alternative therapies for the treatment of enteric neuropathies. In stem cell therapy, there are two main techniques. The first approach is to repopulate the aganglionated GI fragments through the transplantation of in vitro derived stem cells. The second method is to steer endogenous stem cells to neurons. Both techniques require the detailed composition of enteric neuronal subtypes and their proportions as well as the required factors involved [5].
Most of the different processes involved in ENS formation, such as migration and proliferation of the ENCCs, has been well studied. The differentiation of neurons and the specification of neuronal subtypes remain largely unknown. To make stem cell therapy possible in the treatment of GI disorders, it is essential to understand each process involved in the ENS formation. This review focuses on the factors and pathways which are shown to be involved in the differentiation and/or subtype specification of enteric neurons (Table 4).
| Factor | Family | Subfamily | Putative roles in the differentiation process |
|---|---|---|---|
| Sox10 | Sox | SoxE | - Keeping ENCCs in undifferentiated state - Differentiation of glial cells - Cooperation with SuFu for ENCC-fate determination |
| Sox6 | Sox | SoxD | - Correlated with gastric dopaminergic neurons - Normal gastric motility |
| Phox2b | Paired homeobox | NA | - Activation of and interaction with Ret in ENCCs - Development of serotonergic and dopaminergic enteric neurons |
| Ret | NA | NA | - Interaction with other factors (Phox2b, Gdnf) |
| Gdnf | Gdnf | NA | - Ret activation in ENCCs - Development of myenteric neurons, with the exception of NOS+-neurons |
| Neurturin | Gdnf | NA | - Attracting axons of excitatory motor neurons and/or promoting axon growth and branching of excitatory motor neurons |
| Hh | NA | NA | - Promoting progression of bipotent ENCCs to differentiate into more mature cell states |
| Ascl1 | bHLH | Ascl | - Development of catecholaminergic immature neurons - Development of serotonergic neurons - Timely initiation of neuron differentiation - Stomach → development VIP, TH and Calb-neurons - Intestines → development TH and Calb-neurons |
| Hand2 | bHLH | NA | - Terminal differentiation of TH, NOS and VIP-neurons |
| Et-3/Ednrb | NA | NA | - Inhibition neuronal differentiation rate - Subtype specification |
| Bmp2 | Tgfβ | Bmp | - Induction development nitrergic and catecholaminergic neurons - Opposite effects on early-born versus late-born neuronal phenotypes - Development of nNOS-neurons - Neuron differentiation |
| Zeb2 | Zinc finger E-box binding proteins | NA | - Sox10 and Et-3-interaction - Glial cell differentiation - Inhibition neuronal differentiation |
| RA | NA | NA | - Promoting neuronal differentiation |
Abbreviations: Sox: SRY-like HMG-box; bHLH: Basic Helix-Loop-Helix; Tgfβ: Transforming growth factor β; Bmp: Bone morphogenic protein; Sox10: SRY-like HMG-box 10; Sox6: SRY-like HMG-box 6; Phox2b: Paired-like homeobox 2b; Ret: Rearranged during transfection; Gdnf: Glial cell line-derived neurotrophic factor; Hh: Hedgehog; Ascl1: Achaete-scute family transcription factor 1; Hand2: Heart- and neural crest- derivatives-expressed transcript 2; Et-3: Endothelin-3; Ednrb: Endothelin receptor b; Bmp2: Bone morphogenic protein 2; Zeb2: Zinc finger E-box binding protein 2; RA: Retinoic Acid; ENCCs: Enteric Neural Crest Cells; SuFu: Suppressor of Fused; (n)NOS: (neuronal) Nitric Oxide Synthase; VIP: Vasoactive Intestinal Polypeptide; TH: Tyrosine Hydroxylase; Calb: Calbindin; NA: Non-Applicable
Table 4:Summary of the known factors involved in the differentiation of enteric neurons and their putative functions.
Factors and Pathways Involved in Neuron Differentiation and Enteric Neuronal Specification
For each neural stem cell or progenitor cell, it needs to be decided whether it will take on an enteric neuronal cell fate or a glial cell fate. Though, the neural stem cells initially take on neuronal precursor characteristics, it is only later during development that glial precursors are formed. During development, it is possible for the outcome of neuronal progenitors to change. In some parts of the GI tract, it has been shown that certain factors are involved first in the early ENS development, and later on also in events such as neuronal differentiation, specification and migration [11].
Since the generation of neuronal diversity is defined by multiple combinatorial codes, multiple factors cooperate to produce different neuronal subtypes. Therefore, the factors involved have their individual functions, but they are still prone to be altered, enhanced, or suppressed when put together with other factors (Figure 1).
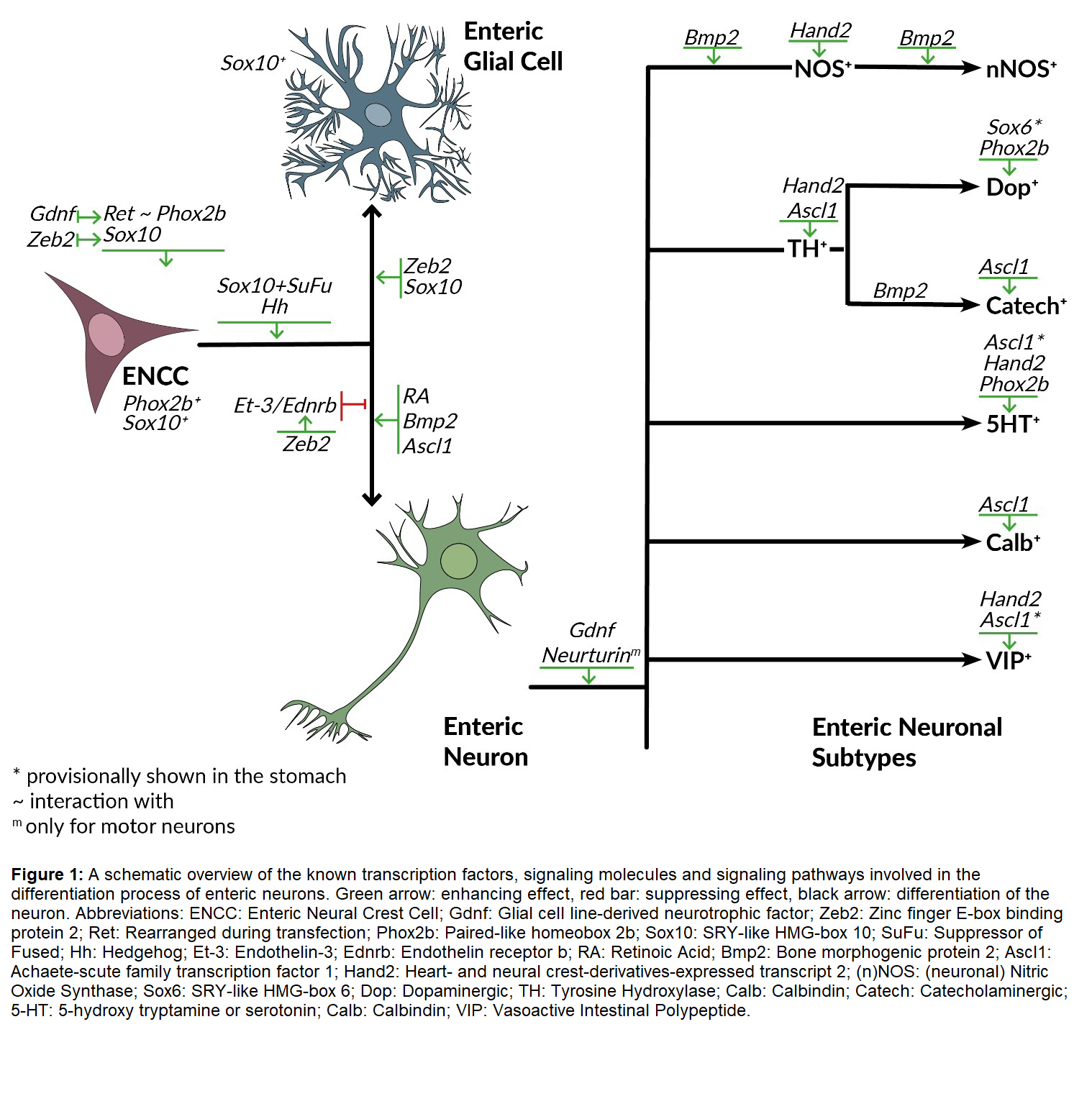
In the following part, the most studied and best-known transcription factors, signaling molecules and pathways, indicated by previous studies to be involved in ENS differentiation are described.
Transcription factors
SRY-like high-mobility group (HMG)-box (Sox) family: This family consists of a group of transcription factors which is characterized by the presence of an HMG domain which has >50% homology with that of the sex-determined-region on the Y chromosome [34]. The proteins in this family are known to affect stem cell functioning and fate through the regulation of expression genes involved in self-renewal and multipotency. It binds to the minor groove of DNA, has a preference for the sequence A/TAACAAT, and triggers conformational changes [34-36]. In the genome of vertebrates, there are around 20 Sox family members which have a broad range of developmental functions, and which are classified in subfamilies going from A until J according to the homology in sequence with those of the Drosophila melanogaster and Caenorhabditis elegans [34,35,37]. The SoxD subfamily, which contains Sox6 transcription factor, is characterized by a long and evolutionary conserved N-terminal domain consisting of a leucine zipper, two coiled-coil domains and a glutamine-rich motif [38]. SoxD genes contain multiple exons in the genomic transcripts, due to which they are alternatively transcribed into long or short transcripts at different stages during development. This enables the genes to fulfil multiple functions [38,39]. Sox10 belongs to the SoxE subfamily which is characterized by 2 domains: an unique dimerization domain proximal to the N-terminus of HMG box, and a distinct C-terminal transactivation domain [39,40]. Sox proteins contain nucleocytoplasmatic shuttling properties which causes their cellular events to be dynamic and complex [41]. The partner protein recruitment and interactions performed by Sox proteins are extensively reviewed by She and Yang in 2015 [39]. In this family, there are also members expressed in neuronal progenitors, such as Sox2, 8 and 10, of which Sox10 is the best-known. Recently, five other Sox family members are found to be involved in the ENS formation. The expression of Sox5 largely coincided with progenitors; Sox4, Sox9 and Sox11 were detected in neurons, and Sox6-expression appeared in neuron subpopulations. Sox4, 9 and 11 are thought to complement the currently known set of Sox genes in the sequential differentiation process of ENS neurons [25].
Sox10: The transcription factor Sox10 belongs to the SoxE subfamily and is known to be essential for the survival and maintenance of undifferentiated ENCCs and glial differentiation [30,35,42]. Its expression is regulated by other transcription factors and by two enhancing domains: one domain crucial for the initiation of Sox10 in newly formed NCs (Sox10E2), and a second domain which plays a role later during development in migrating truncal and vagal crest cells (Sox10E1) [35]. Sox10 is expressed early in development in all delaminating NCCs and later in the ENCCs [42,43]. Therefore, it is often used as an NCC-marker. However, Sox10 is not required for the formation of NCCs since the generation and the emigration of NCCs are unaltered in homozygous (Sox10-/-) mutants. On the other hand, forced expression has been suggested to impose NCC-like characteristics in in vivo neural tissue [42]. The expression of Sox10 will be downregulated as cells differentiate into enteric neurons [11,20,44]. While the Sox10-expression is decreasing in certain cells, in some other cells the expression remains. These cells are either maintaining the undifferentiated neuronal state, or becoming glial cells [11,43,44]. The regulation of Sox10-expression levels determines the differentiation between neuronal and glial lineages, and forms thus an important factor in the ENS development [30]. A role in the early survival of ENCCs has been suggested through mouse models lacking Sox10. In these mutants, the NCCs fail to colonize the largest portion of the GI tract [11]. This failing is a result of extensive cell death of the vagal NCCs prior to the colonization process. This leads to an absence of ENS in this mouse model. On the other hand, an overexpression of Sox10 does not seem to have an effect on the commitment of ENCCs to the neurogenic lineage [42]. In addition, heterozygous Sox10 mutants displayed a reduced number of progenitor cells at early stages of the ENS development [45].
Sox6: Sox6 is a member of the SoxD subfamily which recently has been identified. Sox6 initiates its expression in mice on E11.5 in a small subset of Sox10+-progenitors, which is later during development than Sox10. The expression then gradually increases until E15.5, and eventually coincides with mature neurons characterized by HuC/D-expression. In addition, it is found that Sox6 co-localizes with gastric tyrosine hydroxylase (TH+) -expressing (dopaminergic) neurons, and that it is also expressed in a gastric subpopulation characterized by neuropeptide Y/calbindin 1 (NPY/Calb1)-expression. These findings led to the conclusion that Sox6 correlates with the formation of gastric dopaminergic neurons. Through gene knockout it was suggested that Sox6 has a function in the generation of normal gastric motility [25].
Paired-like homeobox 2b (Phox2b): Phox2b is a homeodomain transcription factor which is initially not expressed in NCCs, but will be as soon progenitor cells enter the enteric mesenchyme [46,47]. Phox2b expression is then observed in enteric neuronal progenitor cells as well as in differentiating neurons during ENS development [43]. This delayed expression of Phox2b was observed in zebrafish in which at 60 hours post fertilization, Phox2b-expression presented the same expression pattern as Sox10, i.e., two parallel rows lateral in the developing GI tract. Since the Phox2b-expression was not as extended as the Sox10- expression, it was suggested that expression of Phox2b starts later than Sox10-expression [48]. Phox2b is seen as a main regulator of neuronal subtype specification. As Sox10, Phox2b is first expressed in enteric precursors, but unlike the Sox10-expression which decreases over time in differentiating enteric neurons, the expression of Phox2b remains [43,46,49]. This balance that exists between Sox10 and Phox2b is demonstrated to be the result of their mutual and reciprocal suppression. When a mutation occurs in Phox2b, it disturbs the balance which leads to the inhibition of early proliferation and subsequentially causes biased differentiation of progenitors towards the glial cell fate. In addition, such a mutation also reduces early proliferation of precursors in enteric glia primordia. It is known that continued proliferation of precursors is of extreme importance in the GI tract since the colonization of the complete GI tissue depends on it. Therefore, mutations in Phox2b are partially responsible for the terminal aganglionosis of HSCR [46].
An important role during development, is the genetic interaction with and activation of the receptor tyrosine kinase rearranged during transfection (Ret) in the progenitor cells [43,46,49]. Besides the regulation of Ret-expression, Phox2b also plays an important role in the development of serotonergic and dopaminergic enteric neurons [47]. In human tissue, it has also been shown that Phox2b is expressed in myenteric neurons of the adult colon. However, not all enteric neurons express Phox2b and even some enteric neurons have the Phox2b expression restricted to neuronal somata. The physiological significance of this variable cellular expression still needs to be revealed [49].
Basic helix-loop-helix (bHLH) family: The transcription factors belonging to this family have conserved a domain comprising a short stretch of basic amino acids and two α helices, separated by a loop motif. It was found that these transcription factors play roles in the regulation of cell-fate determination and the development of certain organs. These factors are divided into different groups depending on either function or expression [50].
Ascl1: Ascl1, initially called Mash1, is a member of the bHLH family belonging to the class B subfamily of the classification based upon expression patterns, and is a transcription factor expressed in both the CNS and the PNS. The onset of Ascl1- expression in ENCCs is induced by a signaling molecule, namely the bone morphogenic protein 2 (Bmp2), and is correlated with the entry of the NCCs in the foregut which occurs early during development [44,50,51]. Along the development, Ascl1 was initially confined with the Sox10+- cell population which are the enteric neuronal progenitors. Then, Ascl1-expression became visible in a subset of HuC/Dexpressing cells at early stages. This subset of neurons also expressed Sox10 and the cell cycle marker Ki67. This showed that these cells were immature neurons. Further during development, it was seen that Ascl1-expression reduced until it remained present in about 5% of the HuC/D-expressing cells. This led to the conclusion that Ascl1 is mainly expressed in ENCCs (progenitors of all neuronal subtypes), after which the expression is maintained for a short period of time for neuronal subtype specification. This expression pattern is found to be conserved in higher vertebrates [51].
Mice lacking Ascl1 showed a decrease of 80% in the number of catecholaminergic immature neurons between E10-14. At E18.5-19 a reduction of HuC/D-expressing cells along the entire GI tract was observed compared to wildtype populations. The reduction is the strongest in the anterior part of the GI tract, and gradually normalizes when going more posterior. This suggests that an upregulation of partially compensating proneural genes is involved. This was tested with the proneural gene Ngn2, which is found to be capable of compensating for Ascl1-loss and induces normal neurogenesis in the ENS without respecifying the ENCCs. This proves that the deletion of Ascl1 leads to a delay in the enteric neurogenesis on one hand, and to the loss of a part of the enteric neuron lineages on the other hand [44,51]. Ascl1 is shown to be involved in the formation of enteric neurons in the esophagus, and in the formation of 5-hydroxytryptamine (5-HT, serotonin) neurons in the intestines and stomach [11,25]. However, not all Ascl1-expressing NCCs are destined to become 5-HT-neurons since around 50% of NCCs express Ascl1, but only 1% of the enteric neurons are 5-HT-neurons. With the downregulation of Sox10 during development, an increase in the Ascl1-expression is seen in a subpopulation of the cells. This Ascl1 upregulation is suggested to suppress the Sox10-expression, therefore Ascl1 is thought to give rise to neurons [11]. It is confirmed that Ascl1-expression is not essential for ENCC-migration and survival, but solely for the timely initiation of neuronal differentiation. It was also found that in mice lacking Ascl1, the percentages of neurons expressing Calb, vasoactive intestinal polypeptide (VIP) or TH were significantly reduced at E18.5-E19 in the stomach. In the intestine, Calb-expressing cells were also strongly reduced, but the number of VIP-expressing cells remained the same. TH-expressing cells were too low to quantify, but a reduction in the intestine was visible. Surprisingly, the number of 5-HTexpressing cells was not affected by the deletion of Ascl1, and even a small, nonsignificant increase was seen. Looking at the expression of calcitonin gene-related peptide (CGRP) and NPY, no difference in numbers of neurons was observed. For NOS+- cells, a slight but also nonsignificant increase was detected. As already quoted, Ngn2 may serve as a compensator for the loss of Ascl1, nevertheless the production of neurons expressing Calb, VIP (stomach) and TH (stomach) remained drastically reduced in these mice, proving the important role of Ascl1 in the formation of specific neuronal subtypes [51].
Heart- and neural crest derivatives-expressed transcript 2 (Hand2): Hand2 is another transcription factor belonging to the bHLH family. It is expressed in nearly all lineages of the ANS, except for the sensory ganglia. In the forming of sympathetic ganglia, the expression of Hand2 is initiated when the precursors were directed towards a sympathetic neuronal cell fate, and already showed Phox2b-expression [52]. This shows the importance of this transcription factor later during ENS development. When it comes to the function of Hand2 in the ANS, there are multiple loss-of-function studies performed in zebrafish and mice indicating its importance in neuroblast differentiation and proliferation [53-55]. Also, the deletion of Hand2 in mouse-NCCs resulted in a reduction of the proliferation of cells expressing the pan-neuronal markers HuC/D and TuJ1 [55]. Another study demonstrated that Hand2 is important for the induction of another transcription factor: Tbx3 [56].
For the ENS, it is shown that in Hand2-/- mice models, the cells express undifferentiated cell markers, such as Sox10, p75, Ret and Phox2b, as well as glial precursors. Only a small amount of pan-neuronal markers was found. In addition, no neuronal subtype-specific markers, such as TH (ANS), NOS and VIP (ENS) were expressed. This indicates that Hand2 is required for terminal differentiation of enteric neurons, and is not involved in the development of glial cells [11,55,56].
Zinc finger E-box binding protein 2 (Zeb2): Zeb2 is a member of the small Zeb family which consists of two members: Zeb1 (δ EF1) and Zeb2 (Sip1 or Zfhx1b). These proteins are characterized by the presence of a homeodomain separated by two clusters of “zinc finger” (C2H2-type) domains which have DNA-binding activity [57,58]. The two transcription factors in this family are characterized by being able to repress transcription by directly binding with each of their two clusters to a series of separated E-box-like sequences (CACCT(G) or CACANNT(G)) in the 5’ regulatory regions of the target genes. Zeb proteins are thought to achieve repression by competing with bHLH transcriptional activator at gene promotors since E-box-like sequences overlap with the target DNA-binding sites of the transcription factors of the bHLH-family [58].
During embryogenesis, Zeb2 is expressed in multiple tissues. Later, it is highly expressed in different neuronal subtypes in the CNS [57]. For the formation of the ENS, Zeb2 is found the be important early during development and is expressed in enteric neuronal progenitors. In addition, the interaction between Zeb2 and Sox10 is essential for the proliferation and differentiation of enteric neurons. Throughout the development, Zeb2-expression is maintained in glial cells but is turned off in mature enteric neurons [59]. The continuous expression of Zeb2 throughout the development highlights its important regulatory role. This has been proven by the observed severe neurological consequences as a result of the loss of Zeb2-expression [58]. Zeb2 knockout studies in mice have also shown severe defects in neural crest formation at E8.5, and death at E9.5 [58,60]. In addition, the deletion of Zeb2 (Zeb2-/-) results in a complete lack of vagal NCCs [59,60]. In heterozygous Zeb2-models, Zeb2+/-, there is an increase in neuronal differentiation which results in more TuJ1+-cells with visible longer axons. The number of neurons which was significantly increased, came together with a significant reduction of Sox10+-cells. The number of glial cells remained the same. This shows that the reduction of Sox10-expression was correlated with a reduction of enteric neuronal progenitors being a consequence of the increase in neuronal differentiation. This indicates some sort of cooperation between Sox10 and Zeb2 [59,61]. Zeb2 also seems to influence the neurogenetic inhibition induced by Et-3. In Zeb2-heterozygous models, when being exposed to Et-3, the inhibition of neurogenesis was only partial. This suggests that the response to Et-3 is incomplete in these models. Of this came the conclusion that coordinate activities of the Et-3/Ednrb-signaling pathway and Zeb2 are required for the differentiation of multi-lineage ENS progenitors in vitro. Looking at the ENS formation in double heterozygous models in vivo, it is seen that the colonic region lacked TuJ1-expression. This suggests that the combined Et-3 and Zeb2 gene dosage reduction impacts the ENS development. This might be the consequence of a genetic interaction between the two loci. Double heterozygous models were also formed between Zeb2 and Ednrb. This resulted in the absence of neurons around the cecum area or starting halfway through the colon. Since Ednrb is the Et-3-receptor in enteric cells, it is strongly suggested that Zeb2 is also involved in a genetic interaction with Ednrb. The investigation of the crosstalk between Zeb2 and Et-3/Ednrbsignaling pathway resulted in a significant increase in neuronal differentiation in double heterozygous models (Zeb2/Et-3). This led to the conclusion that a coordinate action between Zeb2 and Et-3 is required to control neuronal differentiation of enteric progenitors in vivo [59].
Homeobox (Hox) and Transcription activator-like effector (Tale): In the developing ENS, four Tale (3 amino acid loop extension) genes were identified: Pbx2, Pbx3, Meis2 and Pknox1. To impose differential transcriptional read-out, the Hox genes are dependent on the interaction with genes of the Tale family. Therefore, it is hypothesized that a combinatorial expression of various Hox and Tale proteins contributes to the presence of subtype-specific characteristics in enteric neurons [25].
Signaling molecules
Bone morphogenic protein 2 (Bmp2): Bmp2 is a signaling factor belonging to the transforming growth factor β (TGFβ) family. Bmp proteins are known to regulate critical functions in embryonic development such as organ morphogenesis and cell migration, proliferation and differentiation within the developing GI tract [62]. Multiple studies reported that Bmp2 plays an important role in ENS development. It is shown to induce differentiation of nitrergic and catecholaminergic neurons, and has a predominant effect on the inhibitory set of enteric neurons without affecting the excitatory neurons [63]. A decrease of Bmp2-signaling also leads to enhanced early-born neuronal phenotypes while the later born neuronal phenotypes are reduced in number. Increase of Bmp2-signaling results in the opposite effect [64]. Another study showed in vitro a higher number of TuJ+-cells in a medium containing more Bmp2 than in controls. Looking at subtype specification, this study showed that the medium in which extra Bmp2 was added, resulted in a higher number of neuronal NOS (nNOS)- expressing enteric neurons compared to the expression in control medium. No difference was observed between the two media in the number of choline acetyltransferase (ChAT)- expressing neurons. The study confirmed these findings by adding noggin, a Bmp-inhibitor, to a third medium. This resulted in a reduction of electrical field stimulation (EFS)- induced relaxation compared to the untreated constructs [65]. All together, these studies prove the importance of Bmp2 in the differentiation of progenitor cells to enteric neurons and subtype formation.
Glial cell line-derived neurotrophic factor (GDNF) family:
Gdnf: Gdnf is the most important member of the Gdnf family involved in ENS formation and neuron specification [11]. Gdnf is expressed in the GI mesenchyme, binds to Gdnf family receptor α1 (GFR1) during early stages of cell migration and proliferation, and activates the Ret tyrosine kinase expressed in ENCCs [66,67]. In addition, it is shown to be essential in the early survival of ENCCs in the small bowel and colon [11,67]. Its function would be to serve as a chemoattractant for ENS precursors [68]. This important role has made it impossible so far to determine its function later in ENS development in vivo since knockout results in unviable animals. In vitro studies, on the other hand, have shown that Gdnf promotes not only survival of enteric neurons in later stages of the development, but also their proliferation, migration, and neuronal differentiation. In heterozygous mice models (Gdnf+/-), there is a reduction of at least 50% of myenteric neurons in the small intestine and colon, but the percentage of nitric oxide synthase (NOS)-expressing cells was the same as in wild-type mice. Thus, Gdnf is not affecting NOS+-neurons [11]. When GFRα1 is inactivated, a significant reduction of neurons in the midgut is observed, while no differences were observed with regard to the glial cells. This suggests that Gdnf-signaling is required for neuronal differentiation [66].
Neurturin: Another member of the Gdnf family is neurturin. Although mice lacking this signaling molecule are viable and seem to have similar numbers of enteric neurons compared to wild-type mice, a decrease is observed in the number of substance P-axons innervating the circular muscle. Therefore, it seems that neurturin is involved in attracting axons of excitatory motor neurons and/or promoting axon growth and branching [11].
It has also been demonstrated that while the development of the ENS is proceeding, the dependency shifts from Gdnf and GFRα1 towards neurturin and GFRα2 for further trophic support. This shift is similar to the switch in trophic factor dependence observed in the parasympathetic nervous system wherein Gdnf is required early in development for proliferation and migration of neuronal precursors, and neurturin becomes an essential factor for the maintenance of neuronal projections in adult animals. Through gene knockout, the same study found that neurturin only has a minimal effect on enteric neuron survival or proliferation before birth [67].
Signaling pathways
Rearranged during transfection (Ret): Ret is a tyrosine kinase which is functionally connected to a family of four receptors. These receptors all specifically bind to one neurotrophic factor, such as glial derived neurotrophic factor (Gdnf). Early during development, Ret seems to be an essential factor for progenitor migration, while during later stages of development, it shown to be more involved in the promotion of neuronal differentiation. In zebrafish, the co-expression of Ret and Sox10 with Phox2bb led to the discovery of three enteric progenitor subpopulations, and it is also demonstrated that a small subpopulation of differentiating enteric neurons express Ret and Phox2bb [43]. In mice, two different monoisoformic alleles of the Ret-locus were generated, Ret9 and Ret51, expressing the two main Ret isoforms (Ret9andRet51). Mouse models homozygous for Ret9showed a normal embryonic and postnatal development, while the models homozygous forRet51were characterized by distal colonic aganglionosis, suggesting that Ret9 is important in normal ENS development [69].Ret51homozygous mice models also represent a reduced number of ENCCs and a delayed migration in the GI tract, proving Ret’s role in ENCC migration and proliferation. Nonetheless, this defect in colonization is corrected shortly afterwards leading to a normal neuronal network after 5 days. This normal network, however, was only observed in the small intestine. In the large intestine, complete migration failed and was only observed in the proximal third of the colon [70].
Hedgehog (Hh)-signaling: Hh-signaling pathway is involved in GI organogenesis and ENS development, and is mediated by the transcription factors Gli1, Gli2, and Gli3 [30,71]. While Gli1 and Gli2 mainly act as activators, Gli3 works as a repressor. Ectopic human Gli1 overexpression in transgenic mice previously resulted in HSCR-like phenotypes, and aberrant activation of Hh-signaling resulted in premature gliogenesis of ENCCs. This suggests that Gli1-activity is important for ENS development and HSCR pathogenesis. Suppressor of Fused (SuFu) reduces Gli transcriptional activity by promoting the formation of Gli3 suppressor activity, and the cytoplasmatic sequestration of Gli activators. Interestingly, Sox10 downregulates SuFu to promote glial differentiation. This indicates a probable cooperation between Sox10 and SuFu/Gli to determine the fate of the NCCs. A mechanistic link between SuFu/Gli and Sox10 has been shown in mice, which coordinates neuronal and glial lineage differentiation, and NCC migration. These observations provided evidence that there is a connection between Gli-mutations and HSCR patients, and show that a perturbed Sox10-SuFu/Gli regulatory nexus leads to HSCR pathogenesis. This study also demonstrated that a deletion of SuFu leads to a spontaneous induction of NCC differentiation to form neurons and glial cells. SuFu is essential to retain directional migration of the ENCCs for the complete innervation of the GI and for the formation of neuronal networks. The loss of SuFu results in random migration of the ENCCs which leads to a delayed migration and to a disorganized ENS. This also causes impaired differentiation and less neurons (hypoganglionosis). It was also found that SuFu negatively regulates Sox10-expression in ENCCs through the control of Gli transcriptional activity, and that SuFu-expression was elevated in Sox10 mutant mice (Sox10N/+) which suggests a bidirectional regulatory loop involved in the control of Sox10 levels [30].
Besides activating Hh by the overexpression of Gli1, it is also possible to activate Hh through knocking out Ptch1, which is a negative regulator of Hh-signaling. It has been suggested that the activation of the Hh-signaling pathway promotes the progression of bipotent ENCCs to differentiate into more mature cell states [71].
Endothelin-3/Endothelin receptor B (Et-3/Ednrb)- signaling: Despite the unclear regulation of enteric neurogenesis so far, it has been shown that Et-3 and Ednrb are involved in populating the distal regions of the GI tract with enteric neurons. Both in vivo as in vitro studies suggest that Et-3 its main function is to inhibit the neuronal differentiation rate of ENCCs, but it has also been suggested that Et-3- signaling impairs the self-renewal of ENS progenitors by promoting their differentiation into myofibroblasts [11,72]. It is also shown that Et-3-signaling is mainly involved in the migration process and that it may influence the phenotype of specific neuron subpopulations since a difference was found in neurotransmitter-expressions in the small intestine of Et-3- deprived adult mice [11]. In this pathway, it was shown that the expression of Ednrb is regulated by Sox10, and that it is activated by Et-3. Once activated, Ednrb causes inhibition of neuronal differentiation in mixed cultures of the ENS [72]. In Zeb2 heterozygous models, the defective response of Ednrb to Et-3 is corrected by virus-mediated production of Ednrb, confirming the negative effect of Zeb2-decrease on the Et-3/Ednrb-signaling pathway. This proves that Ednrb gene expression is not only directly co-regulated by Sox10, but also by Zeb2 [59].
During the colonization of the small intestine, the ENCCs expressing Et-3 are ahead of the ENCCs expressing Ret and Ednrb. This shows that the Et-3/Ednrb-signaling pathway also plays an important role in the development of the ENS in prececal GI segments. Mice models lacking both Et-3 and specific Ret-loci show a much more severe phenotype compared to the mice models lacking only one of the two. This suggests that there is also a strong genetic interaction between the Ret and Et-3 alleles [70].
Retinoic acid (RA)-signaling: RA is an active vitamin A metabolite which is shown to have an important role in the ENS development. It is thought to be involved in the specification of vagal NCCs and more specifically in the proliferation and migration of ENS progenitors [73,74].
It was first shown that an overexpression of RA, or introducing an excess of RA as a treatment, leads to a delayed colonization of the ENS precursors [75]. Next, it was revealed that inactivation of retinaldehyde dehydrogenase 2 (RALDH2/ALDH1a2), the enzyme responsible for RA-induction during embryonic development, leads to GI aganglionosis [76]. The fact that RA increases neuronal differentiation in other neuronal lineages led to the hypothesis that this is also the case for the ENS. It was shown that indeed, RA enhances the neuronal differentiation of enteric precursors. Interestingly, when it comes to neurite outgrow it was seen that the ones originating from the ENS precursors that were maintained in RA, were significantly lower than in cells grown without RA. These results prove a complex effect of RA on ENS development [73]. A study performed on human pluripotent stem cells (hPSC) showed that the timing of RA-induction also influences enteric neuron differentiation. If RA is expressed too early during development, the induction of NCCs from hPSCs is blocked. Late addition of RA changes the axial identity of cells committed to neural crest fate [74].
Other signaling pathways
In 2018, Memic and her colleagues performed an extensive study searching undetermined transcription factors, signaling molecules and pathways. During this study, 9 novel and unreported signaling pathways were determined: fibroblast growth factor (FGF), activin A (Inhba dimer), TGFβ, chemokine (CXCL12), ephrin/Eph, insulin-like growth factor (IGF), fibronectin leucin rich transmembrane protein (FLRT), connective tissue growth factor (CTGF), and midkine (MDK). These signaling pathways were also found to be expressed in different stages during development. While CTGF and most of the others were expressed throughout the complete ENS development, it was seen that FGF1 is expressed during midstages, and that MDK and activin A were expressed from midto late-stages. In addition, novel receptors having incomplete signaling characterization were also identified: leukocyte receptor tyrosine kinase (LTK) and aryl hydrocarbon receptor (AHR). As with the signaling pathways, a spatiotemporal expression was sometimes found. AHR-expression was only found in the colon between E15-E18, whereas FGF receptor 1 (FGFR1) was expressed along the whole GI tract except in the colon. This study gave rise to signaling components who are indicative of 16 cell-cell communication pathways with unexplored roles in the developing ENS [25].
Conclusion
The development of the ENS is a complex event which involves a series of processes. These processes are guided by the presence and interaction of multiple transcription factors, signaling molecules and pathways. For the treatment of GI disorders due to failing of one of these processes, there is a great interest in the application of stem cell therapy rather than the currently used surgical resection followed by anastomosis. But before stem cell therapy is officially introduced as a possible therapy, the processes underlying ENS formation need to be completely unraveled and understood. A lot of research has been performed on the migration and proliferation processes involved in ENS development, but little is known regarding the differentiation process. So far, only a few transcription factors, signaling molecules and pathways are reported to play certain roles in the differentiation process. This review described the factors of which is has been proven to be involved individually as well as through interactions. Nevertheless, this information still remains insufficient to introduce stem cell therapy in the treatment of GI disorders such as HSCR. Therefore, in the future more research into the differentiation and subtype specification of enteric neurons is required.
Conflicts of Interest
No conflicts of interest.
References
2. Burns AJ, Goldstein AM, Newgreen DF, Stamp L, Schäfer K-H, Metzger M, et al. White paper on guidelines concerning enteric nervous system stem cell therapy for enteric neuropathies. Dev Biol. 2016 Sep;417(2):229–51.
3. Costa M. Anatomy and physiology of the enteric nervous system. Gut. 2000 Dec 1;47(90004):15iv–19.
4. Goldstein A, Hofstra R, Burns A. Building a brain in the gut: development of the enteric nervous system: Building a brain in the gut. Clin Genet. 2013 Apr;83(4):307–16.
5. Morarach K, Mikhailova A, Knoflach V, Memic F, Kumar R, Li W, et al. Diversification of molecularly defined myenteric neuron classes revealed by single-cell RNA sequencing. Nat Neurosci. 2021 Jan;24(1):34–46.
6. Timmermans J-P, Adriaensen D, Cornelissen W, Scheuermann DW. Structural Organization and Neuropeptide Distribution in the Mammalian Enteric Nervous System, with Special Attention to Those Components Involved in Mucosal Reflexes. Comp Biochem Physiol A Physiol. 1997 Oct;118(2):331–40.
7. Furness JB. Types of neurons in the enteric nervous system. J Auton Nerv Syst. 2000 Jul;81(1–3):87–96.
8. Heanue TA, Pachnis V. Enteric nervous system development and Hirschsprung’s disease: advances in genetic and stem cell studies. Nat Rev Neurosci. 2007 Jun;8(6):466–79.
9. Sasselli V, Pachnis V, Burns AJ. The enteric nervous system. Dev Biol. 2012 Jun;366(1):64–73.
10. Rao M, Gershon MD. Enteric nervous system development: what could possibly go wrong? Nat Rev Neurosci. 2018 Sep;19(9):552–65.
11. Hao MM, Young HM. Development of enteric neuron diversity. J Cell Mol Med. 2009 Jul;13(7):1193–210.
12. Smolilo DJ, Costa M, Hibberd TJ, Wattchow DA, Spencer NJ. Morphological evidence for novel enteric neuronal circuitry in guinea pig distal colon. J Comp Neurol. 2018 Jul 1;526(10):1662– 72.
13. Hansen MB. The Enteric Nervous System I: Organisation and Classification: ENTERIC NERVOUS SYSTEM. Pharmacol Toxicol. 2003 Mar;92(3):105–13.
14. Brehmer A. Structure of enteric neurons. Adv Anat Embryol Cell Biol. 2006;186:1–91.
15. Uyttebroek L, Shepherd IT, Harrisson F, Hubens G, Blust R, Timmermans J-P, et al. Neurochemical coding of enteric neurons in adult and embryonic zebrafish (Danio rerio). J Comp Neurol. 2010 Nov 1;518(21):4419–38.
16. Uyttebroek L, Shepherd IT, Hubens G, Timmermans J-P, Van Nassauw L. Expression of neuropeptides and anoctamin 1 in the embryonic and adult zebrafish intestine, revealing neuronal subpopulations and ICC-like cells. Cell Tissue Res. 2013 Nov;354(2):355–70.
17. May-Zhang AA, Tycksen E, Southard-Smith AN, Deal KK, Benthal JT, Buehler DP, et al. Combinatorial Transcriptional Profiling of Mouse and Human Enteric Neurons Identifies Shared and Disparate Subtypes In Situ. Gastroenterology. 2021 Feb;160(3):755-770.e26.
18. Serbedzija GN, Burgan S, Fraser SE, Bronner-Fraser M. Vital dye labelling demonstrates a sacral neural crest contribution to the enteric nervous system of chick and mouse embryos. Dev Camb Engl. 1991 Apr;111(4):857–66.
19. Roy-Carson S, Natukunda K, Chou H, Pal N, Farris C, Schneider SQ, et al. Defining the transcriptomic landscape of the developing enteric nervous system and its cellular environment. BMC Genomics. 2017 Dec;18(1):290.
20. Tian J, Zeng C, Tian Z, Lin Y, Wang B, Pan Y, et al. Downregulation of Protein Tyrosine Phosphatase Receptor Type R Accounts for the Progression of Hirschsprung Disease. Front Mol Neurosci. 2019 Apr 10;12:92.
21. Mederer T, Schmitteckert S, Volz J, Martínez C, Röth R, Thumberger T, et al. A complementary study approach unravels novel players in the pathoetiology of Hirschsprung disease. McCallion AS, editor. PLOS Genet. 2020 Nov 5;16(11):e1009106.
22. Barber K, Studer L, Fattahi F. Derivation of enteric neuron lineages from human pluripotent stem cells. Nat Protoc. 2019 Apr;14(4):1261–79.
23. Kapur RP. Colonization of the Murine Hindgut by Sacral Crest-Derived Neural Precursors: Experimental Support for an Evolutionarily Conserved Model. Dev Biol. 2000 Nov;227(1):146– 55.
24. Yu Q, Du M, Zhang W, Liu L, Gao Z, Chen W, et al. Mesenteric Neural Crest Cells Are the Embryological Basis of Skip Segment Hirschsprung’s Disease. Cell Mol Gastroenterol Hepatol. 2021;12(1):1–24.
25. Memic F, Knoflach V, Morarach K, Sadler R, Laranjeira C, Hjerling-Leffler J, et al. Transcription and Signaling Regulators in Developing Neuronal Subtypes of Mouse and Human Enteric Nervous System. Gastroenterology. 2018 Feb;154(3):624–36.
26. Desmet A-S, Cirillo C, Vanden Berghe P. Distinct subcellular localization of the neuronal marker HuC/D reveals hypoxiainduced damage in enteric neurons. Neurogastroenterol Motil. 2014 Aug;26(8):1131–43.
27. Webster W. Embryogenesis of the enteric ganglia in normal mice and in mice that develop congenital aganglionic megacolon. J Embryol Exp Morphol. 1973 Dec;30(3):573–85.
28. Parisi MA, Kapur RP. Genetics of Hirschsprung disease: Curr Opin Pediatr. 2000 Dec;12(6):610–7.
29. Chatterjee S, Nandakumar P, Auer DR, Gabriel SB, Chakravarti A. Gene- and tissue-level interactions in normal gastrointestinal development and Hirschsprung disease. Proc Natl Acad Sci. 2019 Dec 26;116(52):26697–708.
30. Liu JA-J, Lai FP-L, Gui H-S, Sham M-H, Tam PK-H, Garcia- Barcelo M-M, et al. Identification of GLI Mutations in Patients With Hirschsprung Disease That Disrupt Enteric Nervous System Development in Mice. Gastroenterology. 2015 Dec;149(7):1837- 1848.e5.
31. Tilghman JM, Ling AY, Turner TN, Sosa MX, Krumm N, Chatterjee S, et al. Molecular Genetic Anatomy and Risk Profile of Hirschsprung’s Disease. N Engl J Med. 2019 Apr 11;380(15):1421– 32.
32. Bhave S, Arciero E, Baker C, Ho WL, Stavely R, Goldstein AM, et al. Enteric neuronal cell therapy reverses architectural changes in a novel diphtheria toxin-mediated model of colonic aganglionosis. Sci Rep. 2019 Dec;9(1):18756.
33. Alhawaj AF. Stem cell-based therapy for hirschsprung disease, do we have the guts to treat? Gene Ther [Internet]. 2021 Jun 14 [cited 2021 Aug 17]; Available from: http://www.nature.com/ articles/s41434-021-00268-4
34. Chung MIS, Ma ACH, Fung T-K, Leung AYH. Characterization of Sry-related HMG box group F genes in zebrafish hematopoiesis. Exp Hematol. 2011 Oct;39(10):986-998.e5.
35. Gou Y, Zhang T, Xu J. Transcription Factors in Craniofacial Development. In: Current Topics in Developmental Biology [Internet]. Elsevier; 2015 [cited 2021 Dec 2]. p. 377–410. Available from: https://linkinghub.elsevier.com/retrieve/pii/ S0070215315000459
36. Dehshahri A, Biagioni A, Bayat H, Lee EHC, Hashemabadi M, Fekri HS, et al. Editing SOX Genes by CRISPR-Cas: Current Insights and Future Perspectives. Int J Mol Sci. 2021 Oct 20;22(21):11321.
37. Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013 Oct 15;140(20):4129–44.
38. Ikeda T, Zhang J, Chano T, Mabuchi A, Fukuda A, Kawaguchi H, et al. Identification and characterization of the human long form of Sox5 (L-SOX5) gene. Gene. 2002 Sep;298(1):59–68.
39. She Z-Y, Yang W-X. SOX family transcription factors involved in diverse cellular events during development. Eur J Cell Biol. 2015 Dec;94(12):547–63.
40. Bernard P. Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Hum Mol Genet. 2003 Jul 15;12(14):1755–65.
41. Malki S, Boizet-Bonhoure B, Poulat F. Shuttling of SOX proteins. Int J Biochem Cell Biol. 2010 Mar;42(3):411–6.
42. Bondurand N, Sham MH. The role of SOX10 during enteric nervous system development. Dev Biol. 2013 Oct;382(1):330–43.
43. Taylor CR, Montagne WA, Eisen JS, Ganz J. Molecular fingerprinting delineates progenitor populations in the developing zebrafish enteric nervous system: Zebrafish Ens Progenitor Subpopulations. Dev Dyn. 2016 Nov;245(11):1081–96.
44. Okamura Y, Saga Y. Notch signaling is required for the maintenance of enteric neural crest progenitors. Development. 2008 Nov 1;135(21):3555–65.
45. Paratore C. Sox10 haploinsufficiency affects maintenance of progenitor cells in a mouse model of Hirschsprung disease. Hum Mol Genet. 2002 Nov 15;11(24):3075–85.
46. Gershon MD. NPARM in PHOX2B: why some things just should not be expanded. J Clin Invest. 2012 Sep 4;122(9):3056–8.
47. Pattyn A, Morin X, Cremer H, Goridis C, Brunet J-F. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999 May;399(6734):366–70.
48. Elworthy S, Pinto JP, Pettifer A, Cancela ML, Kelsh RN. Phox2b function in the enteric nervous system is conserved in zebrafish and is sox10-dependent. Mech Dev. 2005 May;122(5):659–69.
49. Cossais F, Lange C, Barrenschee M, Möding M, Ebsen M, Vogel I, et al. Altered enteric expression of the homeobox transcription factor Phox2b in patients with diverticular disease. United Eur Gastroenterol J. 2019 Apr;7(3):349–57.
50. Taneja R, editor. BHLH transcription factors in development and disease. First edition. Amsterdam ; Boston: Elsevier/Academic Press; 2014. 381 p. (Current topics in developmental biology).
51. Memic F, Knoflach V, Sadler R, Tegerstedt G, Sundström E, Guillemot F, et al. Ascl1 Is Required for the Development of Specific Neuronal Subtypes in the Enteric Nervous System. J Neurosci. 2016 Apr 13;36(15):4339–50.
52. Stanzel S, Stubbusch J, Pataskar A, Howard MJ, Deller T, Ernsberger U, et al. Distinct roles of hand2 in developing and adult autonomic neurons. Dev Neurobiol. 2016 Oct;76(10):1111–24.
53. Lucas ME, Müller F, Rüdiger R, Henion PD, Rohrer H. The bHLH transcription factor hand2 is essential for noradrenergic differentiation of sympathetic neurons. Development. 2006 Oct 15;133(20):4015–24.
54. Morikawa Y, D’Autréaux F, Gershon MD, Cserjesi P. Hand2 determines the noradrenergic phenotype in the mouse sympathetic nervous system. Dev Biol. 2007 Jul;307(1):114–26.
55. Hendershot TJ, Liu H, Clouthier DE, Shepherd IT, Coppola E, Studer M, et al. Conditional deletion of Hand2 reveals critical functions in neurogenesis and cell type-specific gene expression for development of neural crest-derived noradrenergic sympathetic ganglion neurons. Dev Biol. 2008 Jul;319(2):179–91.
56. Wright CM, Schneider S, Smith-Edwards KM, Mafra F, Leembruggen AJL, Gonzalez MV, et al. scRNA-Seq Reveals New Enteric Nervous System Roles for GDNF, NRTN, and TBX3. Cell Mol Gastroenterol Hepatol. 2021;11(5):1548-1592.e1.
57. Epifanova E, Babaev A, Newman AG, Tarabykin V. Role of Zeb2/ Sip1 in neuronal development. Brain Res. 2019 Feb;1705:24–31.
58. Hegarty SV, Sullivan AM, O’Keeffe GW. Zeb2: A multifunctional regulator of nervous system development. Prog Neurobiol. 2015 Sep;132:81–95.
59. Watanabe Y, Stanchina L, Lecerf L, Gacem N, Conidi A, Baral V, et al. Differentiation of Mouse Enteric Nervous System Progenitor Cells Is Controlled by Endothelin 3 and Requires Regulation of Ednrb by SOX10 and ZEB2. Gastroenterology. 2017 Apr;152(5):1139-1150.e4.
60. Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D, et al. Mice Lacking Zfhx1b, the Gene That Codes for Smad-Interacting Protein-1, Reveal a Role for Multiple Neural Crest Cell Defects in the Etiology of Hirschsprung Disease–Mental Retardation Syndrome. Am J Hum Genet. 2003 Feb;72(2):465–70.
61. Stanchina L, Van de Putte T, Goossens M, Huylebroeck D, Bondurand N. Genetic interaction between Sox10 and Zfhx1b during enteric nervous system development. Dev Biol. 2010 May;341(2):416–28.
62. Huang S, Wang Y, Luo L, Li X, Jin X, Li S, et al. BMP2 Is Related to Hirschsprung’s Disease and Required for Enteric Nervous System Development. Front Cell Neurosci. 2019 Dec 3;13:523.
63. Anitha M, Shahnavaz N, Qayed E, Joseph I, Gossrau G, Mwangi S, et al. BMP2 promotes differentiation of nitrergic and catecholaminergic enteric neurons through a Smad1- dependent pathway. Am J Physiol-Gastrointest Liver Physiol. 2010 Mar;298(3):G375–83.
64. Chalazonitis A, Kessler JA. Pleiotropic effects of the bone morphogenetic proteins on development of the enteric nervous system. Dev Neurobiol. 2012 Jun;72(6):843–56.
65. Rego SL, Raghavan S, Zakhem E, Bitar KN. Enteric neural differentiation in innervated, physiologically functional, smooth muscle constructs is modulated by bone morphogenic protein 2 secreted by sphincteric smooth muscle cells: Enteric neural differentiation with SMC-derived BMP2. J Tissue Eng Regen Med. 2017 Apr;11(4):1251–61.
66. Uesaka T, Nagashimada M, Enomoto H. GDNF Signaling Levels Control Migration and Neuronal Differentiation of Enteric Ganglion Precursors. J Neurosci. 2013 Oct 9;33(41):16372–82.
67. Gianino S, Grider JR, Cresswell J, Enomoto H, Heuckeroth RO. GDNF availability determines enteric neuron number by controlling precursor proliferation. Development. 2003 May 15;130(10):2187–98.
68. Natarajan D, Marcos-Gutierrez C, Pachnis V, de Graaff E. Requirement of signalling by receptor tyrosine kinase RET for the directed migration of enteric nervous system progenitor cells during mammalian embryogenesis. Development. 2002 Nov 15;129(22):5151–60.
69. de Graaff E. Differential activities of the RET tyrosine kinase receptor isoforms during mammalian embryogenesis. Genes Dev. 2001 Sep 15;15(18):2433–44.
70. Barlow A, de Graaff E, Pachnis V. Enteric Nervous System Progenitors Are Coordinately Controlled by the G Protein-Coupled Receptor EDNRB and the Receptor Tyrosine Kinase RET. Neuron. 2003 Dec;40(5):905–16.
71. Lau S-T, Li Z, Pui-Ling Lai F, Nga-Chu Lui K, Li P, Munera JO, et al. Activation of Hedgehog Signaling Promotes Development of Mouse and Human Enteric Neural Crest Cells, Based on Single-Cell Transcriptome Analyses. Gastroenterology. 2019 Dec;157(6):1556- 1571.e5.
72. Bondurand N, Natarajan D, Barlow A, Thapar N, Pachnis V. Maintenance of mammalian enteric nervous system progenitors by SOX10 and endothelin 3 signalling. Development. 2006 May 15;133(10):2075–86.
73. Sato Y, Heuckeroth RO. Retinoic acid regulates murine enteric nervous system precursor proliferation, enhances neuronal precursor differentiation, and reduces neurite growth in vitro. Dev Biol. 2008 Aug;320(1):185–98.
74. Frith TJR, Gogolou A, Hackland JOS, Hewitt ZA, Moore HD, Barbaric I, et al. Retinoic Acid Accelerates the Specification of Enteric Neural Progenitors from In-Vitro-Derived Neural Crest. Stem Cell Rep. 2020 Sep;15(3):557–65.
75. Pitera JE, Smith VV, Woolf AS, Milla PJ. Embryonic Gut Anomalies in a Mouse Model of Retinoic Acid-Induced Caudal Regression Syndrome. Am J Pathol. 2001 Dec;159(6):2321–9.
76. Niederreither K, Vermot J, Roux IL, Schuhbaur B, Chambon P, Dollé P. The regional pattern of retinoic acid synthesis by RALDH2 is essential for the development of posterior pharyngeal arches and the enteric nervous system. Development. 2003 Jun 1;130(11):2525–34.
