Abstract
Innate immunity is the oldest form in evolution and is present in all multicellular organisms, including vertebrates and invertebrates. Although humans are the most recent evolutionary phylum, there is abundant evidence of a genetic inheritance shared between invertebrates and humans. There is correspondence between molecular pathways associated with the recognition systems of pathogen-associated molecular patterns (PAMPs) via pattern recognition receptors (PRRs) and Peptidoglycan recognition proteins (PGRPs). From a molecular point of view, intimate associations occur in key components of the molecular signaling process such as signal transducer and activator of transcription (STAT), Janus kinase (JAK), c-Jun N-terminal kinase (JNK), Toll-like receptor (TLR). Using these parallels is essential for a better understanding and conservation of living organisms, establishing essential biotechnological strategies for the progress of the understanding of the immune system.
Keywords
Innate immunity, Invertebrates, Humans
Abbreviation
PRRs: Pattern Recognition Receptors; PAMPs: Pathogen-Associated Molecular Patterns; TLRs: Toll-Like Receptors; NLRs: Nucleotide-Binding Oligomerization Domain Receptors; ROS: Reactive Oxygen Species; PGRPs: Peptidoglycan Recognition Proteins; AMPs: Antimicrobial Peptides
Introduction
The immune system has multiple pathways of action, and these pathways are divided into two major blocks: innate immunity, which is intrinsic to animals, and adaptive immunity. Innate immunity represents the first line of defense against pathogens in animals and it is characterized by its nonspecific nature. This form of immunity is the oldest form in evolution and is present in all multicellular organisms, including vertebrates and invertebrates [1]. Unlike adaptive immunity, which depends on specific recognition of pathogens and involves a memory component, innate immunity acts immediately after infection and does not require prior exposure to a pathogen, being a more immediate response pathway than adaptive immunity [2].
Innate immune system employs a variety of mechanisms to identify and respond to pathogens. At the heart of this process are pattern recognition receptors (PRRs), which recognize conserved molecular patterns associated with pathogens, known as pathogen-associated molecular patterns (PAMPs) [3]. These receptors include Toll-like receptors (TLRs), which play a critical role in initiating immune responses by detecting microbial components and activating signaling pathways that lead to the production of inflammatory cytokines and antimicrobial peptides (AMPs) [4]. Innate immune responses can be modulated by several factors, including environmental cues and the presence of other immune cells, which can either enhance or suppress the overall immune response [5].
With studies being conducted modulating the action of innate immunity, the concept of “learned immunity” has been highlighted, focusing mainly on clades of animals that only have innate immunity as a form of immune response. Learned immunity occurs when innate immune cells exhibit enhanced responses after re-exposure to pathogens, suggesting a form of immunological memory that was previously considered exclusive to adaptive immunity [6]. Studies such as these are able to highlight the need to better understand how innate immunity works in the body and in animal cells. For this reason, the present work aims to highlight the importance of innate immunity through a review of what is known about innate immunity as a form of response within the evolution of taxa, addressing the theme from more basal animals to mammals.
The Foundation of Immunity: Basal Animals as Evolutionary Evidence
Innate immunity in basal animals, such as poriferans (sponges) and cnidarians (i.e. sea anemones and corals), represents a fundamental aspect of their biological defense mechanisms against pathogens. This first form of defense is characterized by its reliance on germline-encoded receptors and immediate responses to microbial threats, distinguishing it from the adaptive immune responses seen in more complex organisms.
In poriferans, innate immunity is primarily mediated by PRRs, which recognize PAMPs. Previous studies have shown that sponges express TLRs, which play a crucial role in initiating immune responses by triggering signaling pathways that lead to the production of AMPs and inflammatory cytokines [7,8]. The sponge Amphimedon queenslandica [9] has been highlighted for its complex repertoire of nucleotide-binding oligomerization domain receptors (NLRs), which are integral to the innate immune response and can initiate pyroptosis and apoptosis in response to infections [10]. This indicates that, even at this early stage of metazoan evolution, sponges possess sophisticated mechanisms for detecting and responding to pathogens.
Furthermore, sponges already utilize several immune signaling pathways, including MyD88-dependent pathways, which are essential for their defense against bacterial infections [8], and these receptors can be induced upon exposure to microbe-associated molecular patterns, demonstrating their ability to recognize and respond to environmental microbial threats [11]. With evolution, it is possible to see some gains but also similarities between animals, with the innate immunity of Cnidaria being similarly characterized by the presence of TLRs and other immune receptors that facilitate the detection of pathogens. The TLR in sea anemones is involved not only in the detection of pathogens, but also in developmental processes, indicating a dual role in both immunity and growth [12].
Innate immune responses in cnidarians are also marked by the activation of signaling pathways that lead to the expression of immune-related genes, which help in the fight against infections [13]. The evolutionary conservation of innate immune genes among these basal taxa highlights the fundamental nature of these immune mechanisms in early-diverging metazoans [14]. For cnidarians, there is also an important relationship that highlights the effectiveness of their innate immunity, indicating that cnidarians have a diverse repertoire of PRRs, which allows them to recognize both microbial threats and their symbiotic partners, such as photosynthetic microalgae that play an important role in obtaining energy for corals (i.e. zooxanthellae) [3]. This immunological specificity is particularly important to maintain the balance between tolerance and defense, as the host must distinguish between beneficial symbionts and harmful pathogens [15]. Innate immune responses in corals can be modulated to support the symbiotic relationship with zooxanthellae, while providing defense against potential pathogens [16].
In taxa lacking an adaptive immune system, the presence of innate immunity in basal metazoans (such as cnidarians and poriferans) provides essential defense mechanisms that enable these organisms to survive despite environmental stressors. In corals, innate immunity plays a pivotal role in maintaining health by regulating the balance between beneficial and harmful microbial communities. Environmental changes, including rising temperatures and salinity fluctuations driven by climate change, can disrupt this balance, leading to a shift from symbiotic to pathogenic microbiota. This dysbiosis can compromise the health and resilience of Cnidaria. However, the innate immune system contributes to the taxon’s ability to manage such microbial shifts, offering a degree of protection against disease and environmental perturbations [17].
The Role of Modern Invertebrates
Similar to poriferans and cnidarians, molluscs have a robust innate immune system, with specialized hemocytes that play a crucial role in defense against pathogens in both freshwater and marine environments. Hemocytes are the main immune cells in molluscs, analogous to mammalian phagocytes such as neutrophils and macrophages. They are involved in several immunological functions, including phagocytosis, encapsulation, and release of antimicrobial substances [18,19]. These cells circulate in the hemolymph of molluscs of all classes and are recruited to sites of infection, where they can recognize and eliminate pathogens through phagocytosis [20,21]. The ability of hemocytes to produce reactive oxygen species (ROS) and other cytotoxic molecules is critical for the destruction of engulfed pathogens [19].
Snails do not have an adaptive immune system, meaning that their immune responses are not specific or anticipatory. Instead, they rely on a diverse array of innate immune mechanisms that include the recognition of PAMPs via PRRs [22]. Peptidoglycan recognition proteins (PGRPs), for example, are important components of the molluscan immune response, facilitating the detection of bacterial infections and triggering downstream immune signaling pathways [23].
The arthropod immune system, particularly in the model organism Drosophila melanogaster, is characterized by a highly efficient innate immune response that plays a crucial role in defense against pathogens, a trait that can be traced back to molluscs. This immune response is mediated primarily by two major signaling pathways: the Toll pathway and the immunodeficiency (Imd) pathway. Both pathways are activated by PRRs that detect PAMPs, leading to the production of AMPs and other immune effectors [24,25]. Upon recognition of PAMPs, such as peptidoglycan from bacterial cell walls or β-glucans from fungal cell walls, the Toll receptor activates a signaling cascade that ultimately leads to the activation of the transcription factor NF-κB (Nuclear factor kappa-light-chain-enhancer of activated B cells), which promotes the expression of several AMPs [26,27].
Recent studies have highlighted the role of specific Toll receptors, such as Toll-7, in the recognition of viral infections and in the activation of autophagy as a defense mechanism [26]. This indicates that the Toll pathway is not only involved in the response to fungal pathogens, but also plays a significant role in antiviral immunity, showing the versatility of this immune signaling pathway. The innate immune mechanisms observed in Drosophila are remarkably conserved among arthropods, indicating a common evolutionary origin. Comparative genomics has revealed that many immune genes are shared among different arthropod species, highlighting the importance of innate immunity in this diverse group [28]. The evolutionary conservation of the Toll and Imd pathways highlights their fundamental roles in host defense against infections.
Finally, among invertebrates, and their last link before they have adaptive immunity, are the Echinoderms. Like arthropods, echinoderms use a variety of cellular and humoral mechanisms to defend themselves against pathogens. Coelomocytes are specialized immune cells that play a crucial role in the innate immune response of echinoderms. They are produced in the axial organ and circulate within the coelomic fluid, where they can respond rapidly to infections or injuries [29,30]. There are several types of coelomocytes, including phagocytes, spherulocytes, and amoebocytes, each with distinct functions in immune defense [31,32]. Phagocytes are particularly important as they are responsible for the process of phagocytosis, where they engulf and digest pathogens such as bacteria and viruses [32,33]. The ability of coelomocytes to recognize and eliminate foreign materials is critical for maintaining the health of echinoderms in their marine environments, as the coelomocytes of the sea cucumber Apostichopus japonicus can effectively phagocytose Vibrio splendidus, a common marine pathogen [32].
AMPs are an essential component of the echinoderm immune system. These peptides are produced by coelomocytes and play a crucial role in the direct targeting and neutralization of pathogens [31,34]. The diversity of AMPs in echinoderms is remarkable, with some species exhibiting a wide range of antimicrobial activities against various pathogens [34]. Furthermore, echinoderms possess a complement-like system that enhances their immune responses. For example, the purple sea urchin Strongylocentrotus purpuratus has been shown to express a complement homologue, SpC3, in its coelomocytes, which is involved in pathogen recognition and elimination [35].
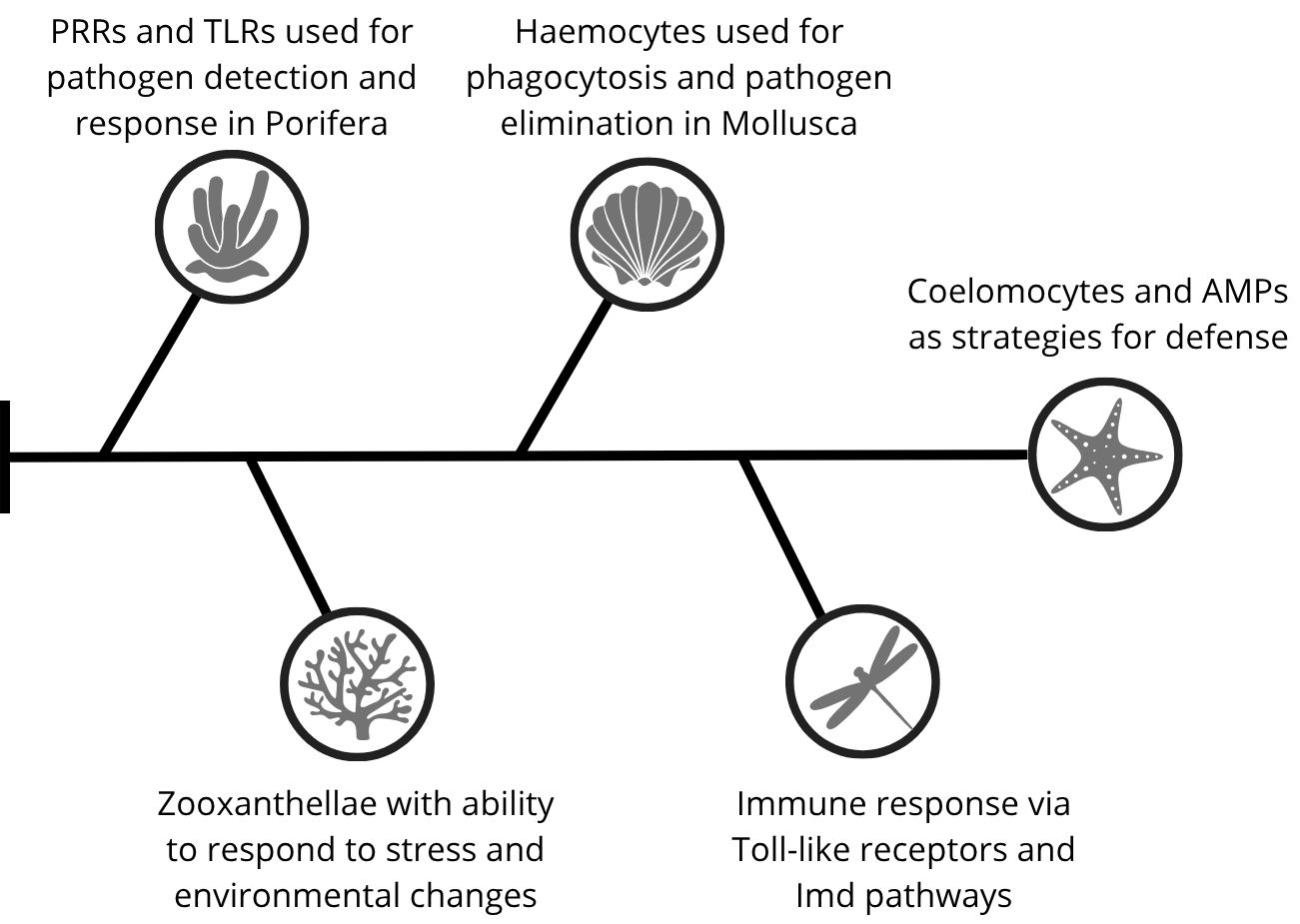
The immune system in invertebrates (Figure 1) carries functions and pathways that can also be seen in vertebrates, and is the precursor to many of the adaptive immunity pathways that vertebrates have evolved to adopt. What can be concluded in relation to these animals is that, although invertebrates do not have adaptive immunity, there are different and common cellular means and functions among all of them that can explain part of how adaptive immunity works and intensified in vertebrate animals.
Figure 1. Evolutionary progression of innate immune components and response strategies in invertebrates. From basal Porifera to the more derived Echinodermata.
Molecular Genetics Implication of Immune System in Invertebrates in Relationship to Humans
Although humans are the most recent evolutionary phylum, there is abundant evidence of a genetic inheritance shared between invertebrates and humans. Similarities are essential for the use of model organisms that aid in the understanding of immunological processes, identifying key components that can be used as a biotechnological tool.
Thinking about homology is an extremely interesting intellectual exercise when one aims at sharing evolutionary inheritance. Examples in this sense can be found for signaling systems that make up the innate immune response, such as members of the Janus kinase (JAK) (hopscotch in Drosophila melanogaster), signal transducer and activator of transcription (STAT) (Stat92E/marelle in Drosophila), c-Jun N-terminal kinase (JNK) (basket in Drosophila melanogaster) and p38 (p38b in Drosophila melanogaster) signaling protein family [36–39]. Porifera have shown intrinsic similarities with the JNK proteins of humans. According to the work of Müller and collaborators [40], there is an important process of selective pressure in the presence of introns in genes related to the JNK gene, and an expansion can be observed in humans. In cnidarians, it is also possible to find homologies related to signaling proteins associated with the aforementioned molecular pathways, such as Wnt, Transforming growth factor beta (TGF-β), and Fibroblast growth factor (FGF) [41].
One of the most intriguing homologies concerns the components of the TLR molecular pathway. The association of the human proteins NF-κB, inhibitor of nuclear factor kappa B (IκB), brain-derived neurotrophic factor (BDNF), interleukin-1 receptor-associated kinase 1 (IRAK-1), IκB kinase (IKK) with the proteins dorsal, cactus, spätzle, pelle and kenny in Drosophila melanogaster has already been described [37,42–47]. Other invertebrates such as poriferans [48], ctenophores [49,50] and echinodermatans [51] show similarities with NK-κB and IκB signaling, as well as IRAK1 [52,53].
In this paper, we can observe that it is possible to establish parallels of important molecular pathways between invertebrates and vertebrates, revealing a broad field of scientific application. Thus, the present work has shown that it is possible and, in addition, it can serve as an important starting point as a biotechnological tool. Thus, we believe that the use of these parallels is essential for a better understanding and conservation of living organisms.
References
2. Woznica A, Kumar A, Sturge CR, Xing C, King N, Pfeiffer JK. STING mediates immune responses in the closest living relatives of animals. Elife. 2021 Nov 3;10:e70436.
3. Emery MA, Dimos BA, Mydlarz LD. Cnidarian Pattern Recognition Receptor Repertoires Reflect Both Phylogeny and Life History Traits. Front Immunol. 2021 Jun 23;12:689463.
4. Richter DJ, Fozouni P, Eisen MB, King N. Gene family innovation, conservation and loss on the animal stem lineage. Elife. 2018 May 31;7:e34226.
5. Netea MG, Joosten LA, Latz E, Mills KH, Natoli G, Stunnenberg HG, et al. Trained immunity: A program of innate immune memory in health and disease. Science. 2016 Apr 22;352(6284):aaf1098.
6. Yoshimura Y, Nii T, Isobe N. Innate Immune Training in Chickens for Improved Defense against Pathogens: A Review. J Poult Sci. 2024 Mar 13;61:2024008.
7. Guzman C, Conaco C. Gene Expression Dynamics Accompanying the Sponge Thermal Stress Response. PLoS One. 2016 Oct 27;11(10):e0165368.
8. Wiens M, Korzhev M, Krasko A, Thakur NL, Perović-Ottstadt S, Breter HJ, et al. Innate immune defense of the sponge Suberites domuncula against bacteria involves a MyD88-dependent signaling pathway. Induction of a perforin-like molecule. J Biol Chem. 2005 Jul 29;280(30):27949–59.
9. Hooper JN, Van Soest RW. A new species of Amphimedon (Porifera, Demospongiae, Haplosclerida, Niphatidae) from the Capricorn-Bunker Group of Islands, Great Barrier Reef, Australia: target species for the ‘sponge genome project’. Zootaxa. 2006 Sep 14;1314(1):31–9.
10. Yuen B, Bayes JM, Degnan SM. The characterization of sponge NLRs provides insight into the origin and evolution of this innate immune gene family in animals. Mol Biol Evol. 2014 Jan 1;31(1):106–20.
11. Pita L, Hoeppner MP, Ribes M, Hentschel U. Differential expression of immune receptors in two marine sponges upon exposure to microbial-associated molecular patterns. Sci Rep. 2018 Oct 31;8(1):16081.
12. Brennan JJ, Messerschmidt JL, Williams LM, Matthews BJ, Reynoso M, Gilmore TD. Sea anemone model has a single Toll-like receptor that can function in pathogen detection, NF-κB signal transduction, and development. Proc Natl Acad Sci U S A. 2017 Nov 21;114(47):E10122–31.
13. van der Burg CA, Prentis PJ, Surm JM, Pavasovic A. Insights into the innate immunome of actiniarians using a comparative genomic approach. BMC Genomics. 2016 Nov 2;17(1):850.
14. Traylor-Knowles N, Browne WE, Mydlarz LD, Palmer CV, Rosental B. Innate Immunity in Early Diverging Metazoans. Front Immunol. 2022 Jan 31;13:816550.
15. Parisi MG, Parrinello D, Stabili L, Cammarata M. Cnidarian immunity and the repertoire of defense mechanisms in anthozoans. Biology. 2020 Sep 11;9(9):283.
16. Stabili L, Parisi MG, Parrinello D, Cammarata M. Cnidarian interaction with microbial communities: from aid to animal's health to rejection responses. Mar Drugs. 2018 Aug 23;16(9):296.
17. Pinnow N, Chibani CM, Güllert S, Weiland-Bräuer N. Microbial community changes correlate with impaired host fitness of Aurelia aurita after environmental challenge. Anim Microbiome. 2023 Sep 21;5(1):45.
18. Allam B, Raftos D. Immune responses to infectious diseases in bivalves. J Invertebr Pathol. 2015 Oct 1;131:121–36.
19. Matozzo V, Chinellato A, Munari M, Finos L, Bressan M, Marin MG. First evidence of immunomodulation in bivalves under seawater acidification and increased temperature. PloS One. 2012 Mar 27;7(3):e33820.
20. Lange MK, Penagos-Tabares F, Muñoz-Caro T, Gärtner U, Mejer H, Schaper R, et al. Gastropod-derived haemocyte extracellular traps entrap metastrongyloid larval stages of Angiostrongylus vasorum, Aelurostrongylus abstrusus and Troglostrongylus brevior. Parasit Vectors. 2017 Jan 31;10(1):50.
21. Al-Khalaifah H, Al-Nasser A. Immune response of molluscs. In: Diarte-Plata G, Escamilla-Montes E, Editors. Molluscs. London: IntechOpen; 2018. P. 1.
22. Rey-Campos M, Moreira R, Gerdol M, Pallavicini A, Novoa B, Figueras A. Immune tolerance in Mytilus galloprovincialis hemocytes after repeated contact with Vibrio splendidus. Front Immunol. 2019 Aug 9;10:1894.
23. Boehm T. Evolution of vertebrate immunity. Curr Biol. 2012 Sep 11;22(17):R722–32.
24. Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. MBio. 2014 Jul 1;5(3):10–128.
25. Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathogens. 2009 Sep 18;5(9):e1000582.
26. Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, et al. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. 2012 Apr 20;36(4):658–67.
27. Imler JL, Tauszig S, Jouanguy E, Forestier C, Hoffmann JA. LPS-induced immune response in Drosophila. J Endotoxin Res. 2000 Dec;6(6):459–62.
28. Palmer WJ, Jiggins FM. Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol Biol Evol. 2015 Aug 1;32(8):2111–29.
29. Mydlarz LD, Jones LE, Harvell CD. Innate immunity, environmental drivers, and disease ecology of marine and freshwater invertebrates. Annu Rev Ecol Evol Syst. 2006 Dec 1;37(1):251–88.
30. Deveci R, Şener E, İzzetoğlu S. Morphological and ultrastructural characterization of sea urchin immune cells. J Morphol. 2015 May;276(5):583–8.
31. Li C, Blencke HM, Haug T, Stensvåg K. Antimicrobial peptides in echinoderm host defense. Dev Comp Immunol. 2015 Mar 1;49(1):190–7.
32. Fu X, Guo M, Liu J, Li C. circRNA432 enhances the coelomocyte phagocytosis via regulating the miR-2008-ELMO1 axis in Vibrio splendidus-challenged Apostichopus japonicus. Commun Biol. 2023 Jan 28;6(1):115.
33. Zhao T, Ren L, Li C, Liu L, Zou Y, Yan H, et al. MiR-7 regulates pathogen-induced immune response via PAK1 in the sea cucumber Apostichopus japonicus. Front Immunol. 2022 Jul 14;13:927796.
34. Solstad RG, Li C, Isaksson J, Johansen J, Svenson J, Stensvåg K, et al. Novel antimicrobial peptides EeCentrocins 1, 2 and EeStrongylocin 2 from the edible sea urchin Echinus esculentus have 6-Br-Trp post-translational modifications. PLoS One. 2016 Mar 23;11(3):e0151820.
35. Gross PS, Clow LA, Smith LC. SpC3, the complement homologue from the purple sea urchin, Strongylocentrotus purpuratus, is expressed in two subpopulations of the phagocytic coelomocytes. Immunogenetics. 2000 Oct;51(12):1034–44.
36. Dearolf CR. JAKs and STATs in invertebrate model organisms. Cellular and Molecular Life Sciences CMLS. 1999 Sep;55(12):1578–84.
37. Kwon EJ, Park HS, Kim YS, Oh EJ, Nishida Y, Matsukage A, et al. Transcriptional Regulation of the Drosophila rafProto-oncogene by Drosophila STAT during Development and in Immune Response. J Biol Chem. 2000 Jun 30;275(26):19824–30.
38. Sluss HK, Han Z, Barrett T, Davis RJ, Ip YT. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 1996 Nov 1;10(21):2745–58.
39. Han ZS, Ip YT. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J Biol Chem. 1999 Jul 23;274(30):21355–61.
40. Mueller SN, Germain RN. Stromal cell contributions to the homeostasis and functionality of the immune system. Nat Rev Immunol. 2009 Sep;9(9):618–29.
41. Matus DQ, Magie CR, Pang K, Martindale MQ, Thomsen GH. The Hedgehog gene family of the cnidarian, Nematostella vectensis, and implications for understanding metazoan Hedgehog pathway evolution. Dev Biol. 2008 Jan 15;313(2):501–18.
42. Ip YT, Reach M, Engstrom Y, Kadalayil L, Cai H, González-Crespo S, et al. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell. 1993 Nov 19;75(4):753–63.
43. Han ZS, Enslen H, Hu X, Meng X, Wu IH, Barrett T, et al. A conserved p38 mitogen-activated protein kinase pathway regulates Drosophila immunity gene expression. Mol Cell Biol. 1998 Jun 1;18(6):3527–39.
44. Hatada EN, Krappmann D, Scheidereit C. NF-κB and the innate immune response. Curr Opin Immunol. 2000 Feb 1;12(1):52–8.
45. Nicolas E, Reichhart JM, Hoffmann JA, Lemaitre B. In vivo regulation of the IκB homologue cactus during the immune response of Drosophila. J Biol Chem. 1998 Apr 24;273(17):10463–9.
46. Mizuguchi K, Parker JS, Blundell TL, Gay NJ. Getting knotted: a model for the structure and activation of Spätzle. Trends Biochem Sci. 1998 Jul 1;23(7):239–42.
47. Kloc M, Halasa M, Kubiak JZ, Ghobrial RM. Invertebrate immunity, natural transplantation immunity, somatic and germ cell parasitism, and transposon defense. Int J Mol Sci. 2024 Jan 16;25(2):1072.
48. Williams LM, Gilmore TD. Looking down on NF-κB. Mol Cell Biol. 2020 Jul 14;40(15):e00104–20.
49. Sinkovics JG. The cnidarian origin of the proto-oncogenes NF-κB/STAT and WNT-like oncogenic pathway drives the ctenophores. Int J Oncol. 2015 Jul 23;47(4):1211–29.
50. Mansfield KM, Carter NM, Nguyen L, Cleves PA, Alshanbayeva A, Williams LM, et al. Transcription factor NF-κB is modulated by symbiotic status in a sea anemone model of cnidarian bleaching. Sci Rep. 2017 Nov 22;7(1):16025.
51. Chovolou Y, Ebada SS, Wätjen W, Proksch P. Identification of angular naphthopyrones from the Philippine echinoderm Comanthus species as inhibitors of the NF-κB signaling pathway. Eur J Pharmacol. 2011 Apr 25;657(1-3):26–34.
52. Emery MA, Dimos BA, Mydlarz LD. Cnidarian pattern recognition receptor repertoires reflect both phylogeny and life history traits. Front Immunol. 2021 Jun 23;12:689463.
53. Gosu V, Basith S, Durai P, Choi S. Molecular evolution and structural features of IRAK family members. PLoS One. 2012 Nov 14;7(11):e49771.