Abstract
The proteasome system in the cell degrades the majority of intracellular proteins. The broad nature of its substrates makes proteasome activity crucial for many cellular functions, such as protein quality control, transcription, apoptosis, immune responses, cell signaling and differentiation. The proteasome system is thus an effective therapeutic target for malignant and non-malignant diseases. In this mini-review, we would like to highlight that proteasome function has also the potential to serve as a biomarker for disease severity and response to treatment. This notion is based on the observation that the six catalytic sites of the proteasome are distinctly altered in peripheral immune cells of patients with chronic inflammatory and autoimmune diseases. We here propose that proteasome activity can be profiled using minimally invasive peripheral blood samples to model severity, progression, and exacerbation of chronic inflammatory diseases. Moreover, the differential alteration of the catalytic activities of the proteasome points towards the existence of multiple catalytic forms of the 20S proteasome that potentially have distinct functions in immune cells. Understanding the role of the distinct immunoproteasome and intermediate proteasome complexes for immune cell function will pave the way for the application of site-specific immunoproteasome inhibitors to specifically target unwanted immune cell functions.
Keywords
Activity-based Probes, Biomarker, Clinical immunology, PBMC, Proteasome
Introduction
The proteasome system is the main protein disposal system in the cell for targeted protein degradation. It mediates the hydrolysis of more than 80% of all cellular proteins, thereby regulating their function in the cell. Degradation products are used to recycle amino acids. A small subset is loaded onto major histocompatibility (MHC) class I molecules to communicate the intracellular protein composition to the immune system [1,2]. Due to the broad nature of substrates, the proteasome is involved in many essential cellular functions, such as protein quality control, transcription, apoptosis, immune responses, cell signaling and differentiation [3,4]. This function makes the proteasome an effective therapeutic target for cancer treatment but also in non-malignant diseases such as cardiovascular, neurodegenerative, lung and autoimmune disorders [5-9].
The proteasome is a multi-subunit protein complex that stands either alone as a 20S catalytic core complex or as a super-complex when bound to one or the other regulator or activator. The most abundant of these super-complexes in the cell is the 26S proteasome, assembled from the 20S core and one or two 19S regulatory particles that attach to one or both ends of the 20S catalytic core, respectively. 26S proteasome complexes enable ubiquitin- and ATP-dependent protein degradation [10]. Other regulators of the proteasome include the proteasome activators (PA) PA28αβ, PA28γ, and PA200, which all mediate ubiquitin-independent substrate degradation [11,12], together with the putative proteasome inhibitor PI31 and proteasome interacting proteins ECM29 or VCP/p97 [13]. Each of these regulators can potentially bind to the 20S core to form singly capped proteasome complexes or doubly capped ones with the potential to assembly into hybrid proteasome complexes that contain two different regulators attached to each end of the 20S proteasome. This gives rise to various different proteasome complexes with the potential to degrade specific proteins at distinct subcellular sites or to modulate proteasome function in a defined manner [13,14].
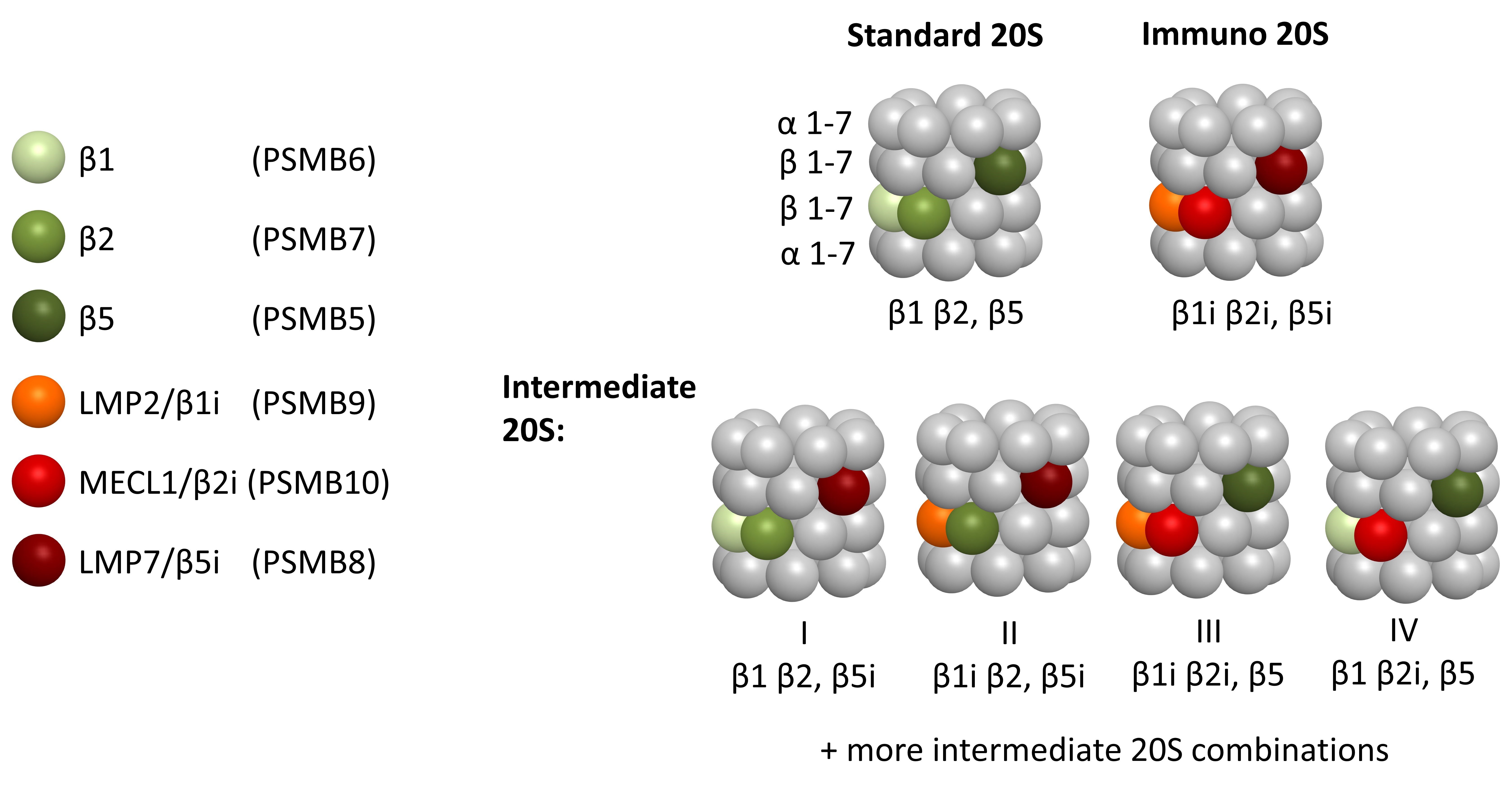
The 20S proteasome consists of a barrel-like hollow structure composed of four stacked heptameric rings (Figure 1); α-subunits form the outer rings (α1-α7), and β-subunits form the inner rings β1-β7 [3,15]. Three of the seven β-subunits of each inner ring are catalytically active and confer the proteolytic capacity of the 20S proteasome that cleave substrates via an N-terminal threonine hydrolase-based mechanism.
The three proteolytic β-subunits determine the species of the 20S core particle: depending on the cell type, cytokine milieu, or activation state of the cell, different β-subunits are expressed and assembled into mature 20S (Figure 1). The standard 20S proteasome is expressed in every cell type and integrates the β1-, β2-, and β5-subunits, which cleave after acidic, basic, or hydrophobic amino acids, respectively, and thus harbor chymotrypsin-like (CT-L), caspase-like (C-L) and trypsin-like (T-L) activities [16]. In immune cells, however, three different β-subunits are constitutively expressed [1]: low molecular mass protein (LMP) 2, multicatalytic endopeptidase complex-like 1 (MECL1), and LMP7 (also called β1i, β2i, and β5i, respectively). In non-immune cells, these three so-called immunosubunits can be induced by IFN-γ or tumor necrosis factor (TNF)-α signaling [17,18] to build the immunoproteasome or induced proteasome. In addition, several other stimuli have been identified that upregulate immunosubunits, including retinoic acid [19], nitric oxide [20], cytokines such as IL-4 [21], Toll-like receptor agonists, type I interferons [22], and recently the mammalian target of rapamycin (mTOR) [23,24]. Importantly, not all immunosubunits are always assembled together to form a full immunoproteasome, but various intermediate proteasomes bearing a mix of immune and standard catalytic subunits exist (Figure 1). Formation of these intermediate complexes might be restricted by the rules of cooperative incorporation of immuno-subunits into nascent and newly assembled proteasomes [25,26]. Accordingly, intermediate complexes containing only a single LMP7/β5i or the two LMP2/β1 and LMP7/β5 immunosubunits (Figure 1, I & II) were identified in various cells and tissues including liver, heart and pancreatic β-cells [27-31]. Other types of intermediate proteasomes (Figure 1, III & IV) were found in experimental systems such as in mice or cells containing a single or combined immunoproteasome knockout. These data suggest that such complexes can be forced to form and that they are stable [17,25,32-37]. Of note, recent data that quantified all proteasome catalytic subunits in human peripheral immune cells using Activity-based Probes (ABPs) also suggested a stoichiometry of intermediate proteasomes other than only single and double immunoproteasomes which conflicts with the reported rules of cooperative incorporation [38,39]. To solve this ambiguity, it is crucial to develop new analytic methods for profiling the activity of each of the proteasome's six distinct catalytic active sites. This will allow for a comprehensive analysis of their cellular function and dysregulation in disease.
Proteasome Activity Profiling Methods
A set of methods have been developed in the last two decades to analyse the different proteasome activities and identify distinct proteasome complexes in a single sample parallel to the analysis of mRNA expression, Mass Spectrometry, ELISA or immunoblotting for protein subunit quantification. The Pros and Cons of each method are summarized in Table 1.
| Proteasome Activity Assay | Pros | Cons |
|---|---|---|
| In-gel proteasome activity Assay |
|
|
| Fluorogenic or luminescent activity assays |
|
|
| Activity-based probes (ABPs) |
|
|
In-gel proteasome activity is used as an approach to dissect the different proteasome complexes, namely the 26S and the 20S proteasome complexes, upon separation on native gels according to their size. With this method, the separated proteasome complexes maintain their activity. They can be quantified by in-gel degradation of a fluorescently quenched substrate, e.g. for the chymotrypsin-like activity of the proteasome. Moreover, blotting of these gels then allows quantification of different proteasome complexes in the gel and to quantify the distinct activators (19S, PA28αβ, PA28γ, PA200, ECM29, VCP/p97) bound to active complexes. When determining both activity and abundance of the different complexes, their ratio determines the specific activity of the distinct proteasome complexes [39-41].
Fluorogenic or luminescent activity assays are in vitro assays to quantify the activity of each β-catalytic site, namely the chymotrypsin-like (CT-L), caspase-like (C-L) and trypsin-like (T-L) proteasome activity, using a specific fluorogenic or luminogenic substrate assay. Substrates consist of catalytic-site specific oligopeptides attached to a quenched reporter (either a fluorogenic or a luminogenic chemical group) which becomes activated upon cleavage. While several of these substrates do not discriminate between standard and immunoproteasomes, recent efforts have led to the development of more specific substrates that are preferentially degraded by either constitutive or immunosubunits [42-45].
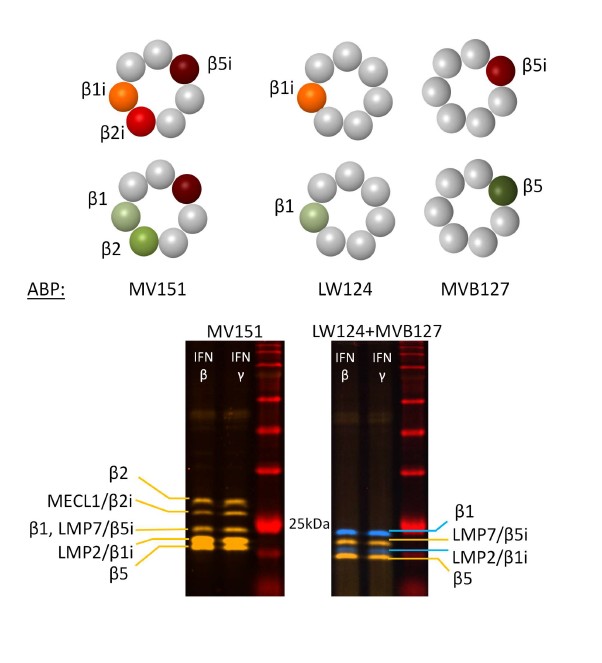
Activity-based probes (ABPs) are another method to quantify the number of active sites in the cell. ABPs bind to the active-site threonine of the respective catalytic subunits and label them covalently with a fluorescent dye. The number of proteolytically active proteasome subunits is quantified by fluorescent detection using two distinct fluorescence channels upon separation of labelled proteasome subunits by SDS-PAGE, enabling identification of the respective subunits according to their molecular weight [46-48] (Figure 2). MV151 is used to label all standard and immuno-subunits of the proteasome and to discriminate the β2 and MECL1 activities, while LW124 and MVB127 specifically bind to the LMP2/β1and LMP7/β5 subunits, respectively (Figure 2) [39,49]. As the ABPs bind to the subunits with the same affinity, the standard versus immune proteasome activities can be directly compared and quantified. With this labelling approach, it is possible to obtain information on the total activity of the proteasome, the activity of the single catalytic subunits, and the ratio of the respective standard versus immune subunits in a single sample. Similar ABPs-based methods are also used to assess the proteasome activity in cell-based imaging procedures [45,50,51]. This profiling methods thereby allows the side-by-side quantification of the different active sites enabling the detection of intermediate proteasome complexes [38].
Standard- and Immunoproteasome Activity Profiles in Health and Disease
Until today, many studies have described dysregulation of the immunoproteasome as a feature of multiple chronic diseases, e.g. neurodegenerative, cardiovascular and age-related diseases, cancer and autoimmune disorders, by quantifying immunosubunit mRNA or protein expression using immunoblotting or ELISA analysis [5,13,52-55]. For example, only LMP7/β5i levels were found to be decreased in the gastrocnemius muscle while only MECL1/β2i was reduced in the tibialis anterior muscle of obese mice as measured by ELISA. Total proteasome in-gel activity in the obese muscles was reduced and inversely correlated with intramuscular accumulation of oxidatively damaged proteins [56]. Another example is human cirrhotic liver samples, where LMP7/β5i was increased both on the mRNA and protein level in response to chronic injury of liver endothelial cells that mediated pro-inflammatory cytokines expression [57]. Similarly, immunoproteasomal mRNA and protein levels were specifically upregulated in pancreatic islets and β-cells in type I diabetic patients and experimental models [29,58], while overall proteasome function was not altered suggesting a pathogenic role for the immunoproteasome [59-61]. This notion was supported experimentally in mice that developed a CD8+ T cell-driven autoimmune form of type I diabetes upon deficiency of MECL1/β2i and LMP7/β5i in their bone-marrow-derived immune cells [62]. As an example of an autoimmune disorder, elevated mRNA levels of only LMP2/β1i were observed in lymphocytes of patients with Sjögren’s Syndrome, pointing to a unique role for this subunit in this disease [63]. High mRNA and protein levels of LMP7/β5i and LMP2/β1i were detected in lymphocytes of patients with myositis [64], and in patients with fast progressive immunoglobulin A nephropathy [65].
As mRNA and protein expression of the proteasome subunits do not necessarily reflect the proteasome subunits’ activity and do not provide any information on the existence of intermediate proteasome complexes, an increasing number of studies are assessing proteasome activities using the methods explained above along with examining proteasomal mRNA and protein expression to determine proteasome regulation in disease. Some studies analyzed blood plasma or bronchoalveolar lavage for an altered amount and activity of extracellular proteasomes as a potential biomarker for acute clinical conditions such as preeclampsia, burn injury, acute ischemic stroke, and acute respiratory stress syndrome [66-69]. Others monitored the blood or bone-marrow-derived samples of patients with multiple myeloma to determine response to treatment and development of resistance to approved proteasome inhibitors [70-72]. These data indicate that proteasome activity profiling is a valuable biomarker to determine response to treatment.
We and others have shown that proteasome activity profile and expression are affected by cigarette smoke and in patients with severe chronic obstructive pulmonary disease (COPD), a major tobacco smoke-related lung disease. While in the lungs, immunoproteasome expression is not altered, its activity is inhibited by cigarette smoke and in COPD [39,73-77]. Of note, immunoproteasome subunits expression was elevated and the activities of 20S and 26S proteasome complexes were altered in Peripheral Blood Mononuclear Cells (PBMCs) of COPD patients [39]. This led us to suggest that proteasome activity profiling in PBMCs might serve as a biomarker for inflammation and pathology severity. We corroborated this concept by examining the proteasome activity profile of PBMCs in the large population-based KORA F4 cohort of 924 subjects [38]. In this study, we demonstrated for the first time, that the proteasome activity profile in PBMCs has a strong sex dimorphism with significantly lower immunoproteasome activity in women. It also changes with age, as almost all catalytic activities of the proteasome were activated in aged women while maintained in ageing men. Moreover, we showed distinct sex-related activation patterns of standard and immunoproteasome active sites in chronic inflammatory diseases such as diabetes, asthma, cardiovascular or chronic obstructive pulmonary diseases [38]. In accordance with these data, a recent study has also suggested profiling the activity of PBMC’s immunoproteasome as a sensitive biomarker for the disease activity of Systemic juvenile idiopathic arthritis (sJIA), enabling the detailed evaluation of response to treatment. This study also demonstrated that proteasome subunits expression, as measured by mass-spectrometry analysis, immediately increased upon inflammatory activation in sJIA [78].
Clinical Potential of Proteasome Activity Profiling as a Biomarker
The high clinical potential of proteasome activity profiling in peripheral blood cells as a biomarker is attributed to the distinct effects each combination of active proteasome subunit has on the cell, as reviewed above. Several studies demonstrated that loss of one of the immunoproteasome subunits changes the cleavage preferences of the 20S proteasome [79] and thereby differentially affects the generation of pathogen-derived MHC class I epitopes, and subsequent pathogen-specific CD8+ T cell responses as reviewed by [1]. Moreover, differential immunoproteasome activity in lymphoid cells impacts their function through intracellular signaling. For instance, immunoproteasome activity, mainly LMP7/β5iactivity, controls the balance of Th17/Treg differentiation through the potentiation of STAT3 activation thereby affecting downstream signaling and cytokine expression [80]. While inhibiting LMP7/β5i activity selectively reduced T cell receptor-induced ERK-phosphorylation in primary T cells, it did not affect NFκB signaling [81]. Of note, neither depletion of MECL1/β1i, LMP2/β2i, or LMP7/β5i impacted canonical NFκB signaling suggesting that the immunoproteasome is not involved in NFκB signaling [82]. Moreover, LMP2/β1i has a major role in the maturation and survival of B and plasma cells and secretion of pro-inflammatory cytokines [35,36,80,81]. In addition, immunoproteasome function controls innate immune responses such as macrophage polarisation and dendritic cell programs [21,33]. Deficiency of LMP7/β5i but not of LMP2/β1i distorts the M2 profile of alveolar macrophages upon IL-4 stimulation [21]. Immunoproteasome activation also emerges as part of a broader response to stress, such as protein and oxidative stress [24,83,84]. Thus, any alteration in immunoproteasome function in lymphoid cells will broadly impact immune cell function, as lymphoid cells express immunoproteasomes or intermediate proteasomes with little standard proteasomes. This proves again that quantifying the distinct activities of the catalytic standard and immune subunits activity will reflect on the immune cell function.
Worth to mention that using proteasome activity as biomarker for disease would also extend to diseases caused by mutations and assembly defects of the proteasome that are increasingly being reported in the literature and that have been grouped into proteasome-associated autoinflammatory syndrome (PRAAS) [14,85]. For example, some mutations in PSMB8 (LMP7/β5) cause impaired assembly of immunoproteasomes resulting in autoinflammation and lipodystrophy, chronic atypical neutrophilic dermatosis, joint contractures, muscle atrophy, microcytic anemia, and autoinflammatory disorders [86-89].
Outlook
Current knowledge indicates differential activities and functions of distinct immunoproteasome complexes in health and disease. Together with the technological advances in activity profiling of the proteasome, this offers an exciting opportunity to apply proteasome activity profiling in peripheral blood cells as a biomarker approach for chronic inflammatory diseases. The use of activity-based probes (ABPs) enables high-throughput profiling and systematic monitoring of proteasome activities in clinical studies using large cohorts of patients or population-based samples, which can then be modelled with clinical data, disease severity and outcome to allow personalized patient stratification. ABP analysis also offers the potential of flow cytometry-based quantification, which can be combined with single subunit expression analysis for defined immune cell populations. In light of the unexpected sex-related regulation of immunoproteasome activity, we urgently need large study cohorts to stratify for sex but also age and BMI [38]. Understanding the role of the distinct immunoproteasome and intermediate proteasome complexes for immune cell function will also pave the way for the application of site-specific immunoproteasome inhibitors to specifically target unwanted immune cell functions, as exemplarily shown for the immunoproteasome specific inhibitor ONX0914 that is currently tested in clinical phase II for Lupus Erythematodes (www.clinicaltrials.gov) [52].
Funding
The work was funded by a personal grant to Silke Meiners by the Leibniz Association (PRO-LUNG).
References
2. Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nature Reviews Immunology. 2003 Dec;3(12):952-61.
3. Ciechanover A, Kwon YT. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Experimental & Molecular Medicine. 2015 Mar;47(3):e147.
4. Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2014 Jan 1;1843(1):13-25.
5. Drews O, Taegtmeyer H. Targeting the ubiquitin-proteasome system in heart disease: the basis for new therapeutic strategies. Antioxidants & Redox Signaling. 2014 Dec 10;21(17):2322-43.
6. Kammerl IE, Meiners S. Proteasome function shapes innate and adaptive immune responses. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2016 Aug 1;311(2):L328-36.
7. Meiners S, Keller IE, Semren N, Caniard A. Regulation of the proteasome: evaluating the lung proteasome as a new therapeutic target. Antioxidants & Redox Signaling. 2014 Dec 10;21(17):2364-82.
8. Morozov AV, Karpov VL. Biological consequences of structural and functional proteasome diversity. Heliyon. 2018 Oct 1;4(10):e00894.
9. Zhang C, Zhu H, Shao J, He R, Xi J, Zhuang R, et al. Immunoproteasome-selective inhibitors: the future of autoimmune diseases?. Future Medicinal Chemistry. 2020 Feb;12(04):269-72.
10. Bard JA, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A. Structure and function of the 26S proteasome. Annual Review of Biochemistry. 2018 Jun 20;87:697.
11. Jiang TX, Zhao M, Qiu XB. Substrate receptors of proteasomes. Biological Reviews. 2018 Nov;93(4):1765-77.
12. Stadtmueller BM, Hill CP. Proteasome activators. Molecular Cell. 2011 Jan 7;41(1):8-19.
13. Barrio R, Sutherland JD, Rodriguez MS, editors. Proteostasis and Disease: From Basic Mechanisms to Clinics. Springer Nature; 2020 Apr 9.
14. Wang X, Meul T, Meiners S. Exploring the proteasome system: a novel concept of proteasome inhibition and regulation. Pharmacology & Therapeutics. 2020 Jul 1;211:107526.
15. Livneh I, Cohen-Kaplan V, Cohen-Rosenzweig C, Avni N, Ciechanover A. The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death. Cell Research. 2016 Aug;26(8):869-85.
16. Huber EM, Basler M, Schwab R, Heinemeyer W, Kirk CJ, Groettrup M, et al. Immuno-and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012 Feb 17;148(4):727-38.
17. Aki M, Shimbara N, Takashina M, Akiyama K, Kagawa S, Tamura T, et al. Interferon-γ induces different subunit organizations and functional diversity of proteasomes. The Journal of Biochemistry. 1994 Feb 1;115(2):257-69.
18. Hallermalm K, Seki K, Wei C, Castelli C, Rivoltini L, Kiessling R, et al. Tumor necrosis factor-α induces coordinated changes in major histocompatibility class I presentation pathway, resulting in increased stability of class I complexes at the cell surface. Blood, The Journal of the American Society of Hematology. 2001 Aug 15;98(4):1108-15.
19. Yang XW, Wang P, Liu JQ, Zhang H, Xi WD, Jia XH, et al. Coordinated regulation of the immunoproteasome subunits by PML/RARα and PU. 1 in acute promyelocytic leukemia. Oncogene. 2014 May;33(21):2700-8.
20. Johnston-Carey HK, Pomatto LC, Davies KJ. The Immunoproteasome in oxidative stress, aging, and disease. Critical Reviews in Biochemistry and Molecular Biology. 2016 Jul 3;51(4):268-81.
21. Chen S, Kammerl IE, Vosyka O, Baumann T, Yu Y, Wu Y, et al. Immunoproteasome dysfunction augments alternative polarization of alveolar macrophages. Cell Death & Differentiation. 2016 Jun;23(6):1026-37.
22. Shin EC, Seifert U, Kato T, Rice CM, Feinstone SM, Kloetzel PM, et al. Virus-induced type I IFN stimulates generation of immunoproteasomes at the site of infection. The Journal of Clinical Investigation. 2006 Nov 1;116(11):3006-14.
23. Choi JH, Jo HS, Lim S, Kim HT, Lee KW, Moon KH, et al. mTORC1 accelerates retinal development via the immunoproteasome. Nature Communications. 2018 Jun 27;9(1):1-6.
24. Yun YS, Kim KH, Tschida B, Sachs Z, Noble-Orcutt KE, Moriarity BS, et al. mTORC1 coordinates protein synthesis and immunoproteasome formation via PRAS40 to prevent accumulation of protein stress. Molecular Cell. 2016 Feb 18;61(4):625-39.
25. Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, et al. Immunoproteasome assembly: cooperative incorporation of interferon γ (IFN-γ)–inducible subunits. The Journal of Experimental Medicine. 1998 Jan 5;187(1):97-104.
26. Groettrup M, Standera S, Stohwasser R, Kloetzel PM. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proceedings of the National Academy of Sciences. 1997 Aug 19;94(17):8970-5.
27. Fabre B, Lambour T, Garrigues L, Ducoux-Petit M, Amalric F, Monsarrat B, et al. Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. Journal of Proteome Research. 2014 Jun 6;13(6):3027-37.
28. Guillaume B, Chapiro J, Stroobant V, Colau D, Van Holle B, Parvizi G, et al. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proceedings of the National Academy of Sciences. 2010 Oct 26;107(43):18599-604.
29. Khilji MS, Verstappen D, Dahlby T, Burstein Prause MC, Pihl C, Bresson SE, et al. The intermediate proteasome is constitutively expressed in pancreatic beta cells and upregulated by stimulatory, low concentrations of interleukin 1 β. PloS one. 2020 Feb 13;15(2):e0222432.
30. Pelletier S, Schuurman KG, Berkers CR, Ovaa H, Heck AJ, Raijmakers R. Quantifying cross-tissue diversity in proteasome complexes by mass spectrometry. Molecular BioSystems. 2010;6(8):1450-3.
31. Abi Habib J, Lesenfants J, Vigneron N, Van den Eynde BJ. Functional differences between proteasome subtypes. Cells. 2022 Jan 26;11(3):421.
32. Dahlmann B. Mammalian proteasome subtypes: Their diversity in structure and function. Archives of Biochemistry and Biophysics. 2016 Feb 1;591:132-40.
33. de Verteuil D, Muratore-Schroeder TL, Granados DP, Fortier MH, Hardy MP, Bramoullé A, et al. Deletion of immunoproteasome subunits imprints on the transcriptome and has a broad impact on peptides presented by major histocompatibility complex I molecules. Molecular & Cellular Proteomics. 2010 Sep 1;9(9):2034-47.
34. Fehling HJ, Swat W, Laplace C, Kühn R, Rajewsky K, Müller U, et al. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science. 1994 Aug 26;265(5176):1234-7.
35. Hensley SE, Zanker D, Dolan BP, David A, Hickman HD, Embry AC, et al. Unexpected role for the immunoproteasome subunit LMP2 in antiviral humoral and innate immune responses. The Journal of Immunology. 2010 Apr 15;184(8):4115-22.
36. Moebius J, van den Broek M, Groettrup M, Basler M. Immunoproteasomes are essential for survival and expansion of T cells in virus‐infected mice. European Journal of Immunology. 2010 Dec;40(12):3439-49.
37. Zanker D, Waithman J, Yewdell JW, Chen W. Mixed proteasomes function to increase viral peptide diversity and broaden antiviral CD8+ T cell responses. The Journal of Immunology. 2013 Jul 1;191(1):52-9.
38. Kammerl IE, Flexeder C, Karrasch S, Thorand B, Heier M, Peters A, et al. Blood immunoproteasome activity is regulated by sex, age and in chronic inflammatory diseases: a first population-based study. Cells. 2021 Nov 28;10(12):3336.
39. Kammerl IE, Hardy S, Flexeder C, Urmann A, Peierl J, Wang Y, et al. Activation of immune cell proteasomes in peripheral blood of smokers and COPD patients: implications for therapy. European Respiratory Journal. 2022 Mar 1;59(3).
40. Odaka H, Ozaki H, Tateno H. scGR-seq: Integrated analysis of glycan and RNA in single cells. STAR protocols. 2022 Mar 18;3(1):101179.
41. Elsasser S, Schmidt M, Finley D. Characterization of the proteasome using native gel electrophoresis. Methods in Enzymology. 2005 Jan 1;398:353-63.
42. Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proceedings of the National Academy of Sciences. 2000 Jul 5;97(14):7754-9.
43. Kim S, Park SH, Choi WH, Lee MJ. Evaluation of Immunoproteasome-Specific Proteolytic Activity Using Fluorogenic Peptide Substrates. Immune Network. 2022 Jun;22(3).
44. Maurits E, Degeling CG, Kisselev AF, Florea BI, Overkleeft HS. Structure‐Based Design of Fluorogenic Substrates Selective for Human Proteasome Subunits. ChemBioChem. 2020 Nov 16;21(22):3220-4.
45. Zerfas BL, Maresh ME, Trader DJ. The immunoproteasome: an emerging target in cancer and autoimmune and neurological disorders. Journal of Medicinal Chemistry. 2019 Oct 31;63(5):1841-58.
46. Iwaya M, Goldman R, Tipper DJ, Feingold B, Strominger JL. Morphology of an Escherichia coli mutant with a temperature-dependent round cell shape. Journal of Bacteriology. 1978 Dec;136(3):1143-58.
47. Hewings DS, Flygare JA, Wertz IE, Bogyo M. Activity‐based probes for the multicatalytic proteasome. The FEBS Journal. 2017 May;284(10):1540-54.
48. Li N, Overkleeft HS, Florea BI. Activity-based protein profiling: an enabling technology in chemical biology Armstrong F, Que Jr L. Current opinion in chemical biology. Current Opinion in Chemical Biology. 2012 Apr;16(1-2):1-2.
49. de Bruin G, Xin BT, Kraus M, van der Stelt M, van der Marel GA, Kisselev AF, et al. A set of activity‐based probes to visualize human (immuno) proteasome activities. Angewandte Chemie. 2016 Mar 18;128(13):4271-5.
50. Carmony KC, Kim KB. Activity-based imaging probes of the proteasome. Cell Biochemistry and Biophysics. 2013 Sep;67(1):91-101.
51. Gan J, Leestemaker Y, Sapmaz A, Ovaa H. Highlighting the proteasome: using fluorescence to visualize proteasome activity and distribution. Frontiers in Molecular Biosciences. 2019 Mar 22;6:14.
52. Basler M, Mundt S, Bitzer A, Schmidt C, Groettrup M. The immunoproteasome: a novel drug target for autoimmune diseases. Clin Exp Rheumatol. 2015 Jul 1;33(4 Suppl 92):S74-9.
53. Basler M, Lindstrom MM, LaStant JJ, Bradshaw JM, Owens TD, Schmidt C, et al. Co‐inhibition of immunoproteasome subunits LMP2 and LMP7 is required to block autoimmunity. EMBO reports. 2018 Dec;19(12):e46512.
54. Keller IE, Vosyka O, Takenaka S, Kloß A, Dahlmann B, Willems LI, et al. Regulation of immunoproteasome function in the lung. Scientific reports. 2015 May 19;5(1):1-2.
55. Tripathi SC, Peters HL, Taguchi A, Katayama H, Wang H, Momin A, et al. Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. Proceedings of the National Academy of Sciences. 2016 Mar 15;113(11):E1555-64.
56. Fletcher E, Wiggs M, Greathouse KL, Morgan G, Gordon PM. Impaired proteostasis in obese skeletal muscle relates to altered immunoproteasome activity. Applied Physiology, Nutrition, and Metabolism. 2022:555-64.
57. Zhang Y, Yang X, Bi T, Wu X, Wang L, Ren Y, et al. Targeted inhibition of the immunoproteasome blocks endothelial MHC class II antigen presentation to CD4+ T cells in chronic liver injury. International Immunopharmacology. 2022 Jun 1;107:108639.
58. Lundh M, Bugliani M, Dahlby T, Chou DH, Wagner B, Ghiasi SM, et al. The immunoproteasome is induced by cytokines and regulates apoptosis in human islets. The Journal of Endocrinology. 2017 Jun;233(3):369.
59. Broca C, Varin E, Armanet M, Tourrel-Cuzin C, Bosco D, Dalle S, et al. Proteasome dysfunction mediates high glucose-induced apoptosis in rodent beta cells and human islets. PloS one. 2014 Mar 18;9(3):e92066.
60. Bugliani M, Liechti R, Cheon H, Suleiman M, Marselli L, Kirkpatrick C, et al. Microarray analysis of isolated human islet transcriptome in type 2 diabetes and the role of the ubiquitin–proteasome system in pancreatic beta cell dysfunction. Molecular and Cellular Endocrinology. 2013 Mar 10;367(1-2):1-0.
61. Casas S, Gomis R, Gribble FM, Altirriba J, Knuutila S, Novials A. Impairment of the ubiquitin-proteasome pathway is a downstream endoplasmic reticulum stress response induced by extracellular human islet amyloid polypeptide and contributes to pancreatic β-cell apoptosis. Diabetes. 2007 Sep 1;56(9):2284-94.
62. Zaiss DM, Bekker CP, Gröne A, Lie BA, Sijts AJ. Proteasome immunosubunits protect against the development of CD8 T cell-mediated autoimmune diseases. The Journal of Immunology. 2011 Sep 1;187(5):2302-9.
63. Krause S, Kuckelkorn U, Dörner T, Burmester GR, Feist E, Kloetzel PM. Immunoproteasome subunit LMP2 expression is deregulated in Sjögren’s syndrome but not in other autoimmune disorders. Annals of the Rheumatic Diseases. 2006 Aug 1;65(8):1021-7.
64. Ghannam K, Martinez-Gamboa L, Spengler L, Krause S, Smiljanovic B, Bonin M, et al. Upregulation of immunoproteasome subunits in myositis indicates active inflammation with involvement of antigen presenting cells, CD8 T-cells and IFNγ. PLoS one. 2014 Aug 6;9(8):e104048.
65. Peruzzi L, Coppo R, Cocchi E, Loiacono E, Bergallo M, Bodria M, et al. The switch from proteasome to immunoproteasome is increased in circulating cells of patients with fast progressive immunoglobulin A nephropathy and associated with defective CD46 expression. Nephrology Dialysis Transplantation. 2021 Aug;36(8):1389-98.
66. Berryman K, Buhimschi CS, Zhao G, Axe M, Locke M, Buhimschi IA. Proteasome levels and activity in pregnancies complicated by severe preeclampsia and hemolysis, elevated liver enzymes, and thrombocytopenia (HELLP) syndrome. Hypertension. 2019 Jun;73(6):1308-18.
67. Chen X, Wang Y, Fu M, Lei H, Cheng Q, Zhang X. Plasma immunoproteasome predicts early hemorrhagic transformation in acute ischemic stroke patients. Journal of Stroke and Cerebrovascular Diseases. 2017 Jan 1;26(1):49-56.
68. Chen XY, Fu M, Wan SF, Zhang X, Wang YZ. Association between plasma immunoproteasome and 90-day prognosis after first-ever ischemic stroke. Neural Regeneration Research. 2021 Apr;16(4):790.
69. Matuszczak E, Weremijewicz A, Komarowska M, Sankiewicz A, Markowska D, Debek W, et al. Immunoproteasome in the plasma of pediatric patients with moderate and major burns, and its correlation with proteasome and UCHL1 measured by SPR imaging biosensors. Journal of Burn Care & Research. 2018 Oct 23;39(6):948-53.
70. Breczko W, Lemancewicz D, Dzięcioł J, Kłoczko J, Bołkun Ł. High immunoproteasome concentration in the plasma of patients with newly diagnosed multiple myeloma treated with bortezomib is predictive of longer OS. Advances in Medical Sciences. 2021 Mar 1;66(1):21-7.
71. Misiewicz-Krzeminska I, de Ramón C, Corchete LA, Krzeminski P, Rojas EA, Isidro I, et al. Quantitative expression of Ikaros, IRF4, and PSMD10 proteins predicts survival in VRD-treated patients with multiple myeloma. Blood advances. 2020 Dec 8;4(23):6023-33.
72. Woodle ES, Tremblay S, Brailey P, Girnita A, Alloway RR, Aronow B, et al. Proteasomal Adaptations Underlying Carfilzomib-Resistance in Human Bone Marrow Plasma Cells. Am J Transplant. 2020;20:399-410.
73. Baker TA, Bach Iv HH, Gamelli RL, Love RB, Majetschak M. Proteasomes in lungs from organ donors and patients with end-stage pulmonary diseases. Physiological Research. 2014 Jun 1;63(3).
74. Kammerl IE, Dann A, Mossina A, Brech D, Lukas C, Vosyka O, et al. Impairment of immunoproteasome function by cigarette smoke and in chronic obstructive pulmonary disease. American Journal of Respiratory and Critical Care Medicine. 2016 Jun 1;193(11):1230-41.
75. Kammerl IE, Caniard A, Merl-Pham J, Ben-Nissan G, Mayr CH, Mossina A, et al. Dissecting the molecular effects of cigarette smoke on proteasome function. Journal of Proteomics. 2019 Feb 20;193:1-9.
76. Somborac‐Bačura A, van der Toorn M, Franciosi L, Slebos DJ, Žanić‐Grubišić T, Bischoff R, et al. Cigarette smoke induces endoplasmic reticulum stress response and proteasomal dysfunction in human alveolar epithelial cells. Experimental Physiology. 2013 Jan;98(1):316-25.
77. van Rijt SH, Keller IE, John G, Kohse K, Yildirim AÖ, Eickelberg O, et al. Acute cigarette smoke exposure impairs proteasome function in the lung. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2012 Nov 1;303(9):L814-23..
78. Sato H, Inoue Y, Kawashima Y, Nakajima D, Ishikawa M, Konno R, et al. In-Depth Serum Proteomics by DIA-MS with In Silico Spectral Libraries Reveals Dynamics during the Active Phase of Systemic Juvenile Idiopathic Arthritis. ACS omega. 2022 Feb 15;7(8):7012-23.
79. Winter MB, La Greca F, Arastu-Kapur S, Caiazza F, Cimermancic P, Buchholz TJ, et al. Immunoproteasome functions explained by divergence in cleavage specificity and regulation. Elife. 2017 Nov 28;6:e27364.
80. Kalim KW, Basler M, Kirk CJ, Groettrup M. Immunoproteasome subunit LMP7 deficiency and inhibition suppresses Th1 and Th17 but enhances regulatory T cell differentiation. The Journal of Immunology. 2012 Oct 15;189(8):4182-93.
81. Schmidt C, Berger T, Groettrup M, Basler M. Immunoproteasome inhibition impairs T and B cell activation by restraining ERK signaling and proteostasis. Frontiers in Immunology. 2018 Oct 26;9:2386.
82. Bitzer A, Basler M, Krappmann D, Groettrup M. Immunoproteasome subunit deficiency has no influence on the canonical pathway of NF-κB activation. Molecular Immunology. 2017 Mar 1;83:147-53.
83. Raynes R, Pomatto LC, Davies KJ. Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Molecular Aspects of Medicine. 2016 Aug 1;50:41-55.
84. Studencka-Turski M, Çetin G, Junker H, Ebstein F, Krüger E. Molecular insight into the IRE1α-mediated type I interferon response induced by proteasome impairment in myeloid cells of the brain. Frontiers in Immunology. 2019 Dec 18;10:2900.
85. Sarrabay G, Méchin D, Salhi A, Boursier G, Rittore C, Crow Y, et al. PSMB10, the last immunoproteasome gene missing for PRAAS. Journal of Allergy and Clinical Immunology. 2020 Mar 1;145(3):1015-7.
86. Treise I, Huber EM, Klein-Rodewald T, Heinemeyer W, Grassmann SA, Basler M, et al. Defective immuno-and thymoproteasome assembly causes severe immunodeficiency. Scientific Reports. 2018 Apr 13;8(1):1-8.
87. Kitamura A, Maekawa Y, Uehara H, Izumi K, Kawachi I, Nishizawa M, et al. A mutation in the immunoproteasome subunit PSMB8 causes autoinflammation and lipodystrophy in humans. The Journal of Clinical Investigation. 2011 Oct 3;121(10):4150-60.
88. Liu Y, Ramot Y, Torrelo A, Paller AS, Si N, Babay S, et al. Mutations in proteasome subunit β type 8 cause chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature with evidence of genetic and phenotypic heterogeneity. Arthritis & Rheumatism. 2012 Mar;64(3):895-907.
89. Agarwal AK, Xing C, DeMartino GN, Mizrachi D, Hernandez MD, Sousa AB, et al. PSMB8 encoding the β5i proteasome subunit is mutated in joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced lipodystrophy syndrome. The American Journal of Human Genetics. 2010 Dec 10;87(6):866-72.
90. Arima K, Kinoshita A, Mishima H, Kanazawa N, Kaneko T, Mizushima T, et al. Proteasome assembly defect due to a proteasome subunit beta type 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo-Nishimura syndrome. Proceedings of the National Academy of Sciences. 2011 Sep 6;108(36):14914-9.