Abstract
Breast cancer remains the world leading cancer type in women. Its management requires a plethora of techniques from imaging to serum analysis; the latter using the presence of mostly protein-based biomarkers for the detection, classification and prediction of breast cancer’s behavior. Saliva is the latest medium in which detection of these biomarkers is being carried out. In this review, we would like to discuss salivary biomarkers for the detection of breast cancer.
Introduction
Breast Cancer is the most regularly diagnosed type of cancer in women in the world, making up on its own 25% of all cases, or nearly 2 million new cases in 2018, and 15% of all cancer related deaths, or around 626,700 deaths for that same year. [1].
The management of breast cancer patients is usually done through biomarker detection, their role is particularly important in deciding the type of systemic therapy for administration [2].
A biomarker is a molecule (protein, nucleic acids, etc) found in either a bodily tissue or fluid that changes its concentration by the presence of one or more types of cancer. It is the product of either by the tumor itself or by the immune response to a tumor. The ideal biomarker has the potential to detect small tumors with great specificity and selectivity in order to either inform for an early diagnosis or help in the tumor’s screening [3].
Biomarker detection technology have also progressed to the point where saliva is now a recognized detection medium that can be collected easily and non-invasively. For the past decade, adjunct tests that use saliva have developed to a level where it plays a key role in improving conventional medical assessments to serious systemic diseases. Hence, the development of a salivary sensitive assay for the accurate, rapid, and non-invasive identification of breast cancer biomarkers would greatly benefit breast cancer’s detection and screening.
When compared to blood, saliva holds several advantages as a diagnostic medium; it is non-invasive and readily available in large quantities, allowing for repeated collection without patient’s discomfort [4].
In addition, salivary biomarkers are a filtrated fraction of their blood counterparts, as such, their analysis offer an advantage through their reflection of the physiological conditions of the body. Analysis of salivary samples can be used to monitor and predict clinical status of systemic diseases [5].
Because of all these advantages, saliva-based detection has been garnering increasing interest and technology based on their detection is offering a promising new clinical strategy for breast cancer [4].
In this mini review, we would like to expand on the salivary protein biomarkers used in breast cancer diagnosis, their biology and their clinical use.
Saliva
Saliva is a biological fluid found in the mouth; it derives from the secretion of the salivary glands in the buccal cavity.
The composition of whole saliva varies depending on a large number of factors and is in itself highly diverse with both body secretions and molecules, and the buccal cavity’s microflora, hence, in recent years, interest has been increasing to develop technologies capable of using saliva as a diagnostic medium [6].
Saliva for diagnosis
The main reason why saliva is garnering that much attention is because of its property of being a filtrated fraction of blood, as such its composition reflects the body’s physiological state. In addition, saliva collection is safer, simpler, and is non-invasive when compared to that of blood. For the diseases where saliva is used for detection, the medium showed great promise and high clinical appeal [5].
Despite that, saliva analysis is considered less reliable than blood or urine analysis; the reason for that is the high difficulty in managing the salivary matrix due to the influence of salivary flow rate on its pH and composition; while also containing lower concentration of said analytes to begin with [7]. In addition, collection methods affect salivary composition greatly, the use of cotton swabs and/or chewing gum to stimulate salivary secretion for example have an effect on decreasing protein concentration while increasing salivary flow rate; while passive drool have the opposite effect that can be attributed to which salivary gland is activated during salivary collection [8].
Saliva and breast cancer
Not many articles have explored the potential of using saliva to detect breast cancer protein biomarkers [1]; the most prominent article regarding this subject was written by Streckfus et al. [9], nearly 20 years ago, it highlighted the correlation of six breast cancer biomarkers found in saliva and their relation to the progression of the disease, these biomarkers will be explained further bellow. However, the small number of participants of this research cast doubt. Since then however, many researchers have focused their attention on Mucin 1 and on Human Epithelial Receptor 2 (HER2) [10-14], while others have focused on other non-protein biomarkers such as microRNA, and metabolomics [4], however, discussing nonprotein antigens is beyond the scope of this review.
Breast Cancer Biomarkers
Epidermal growth factor receptors family
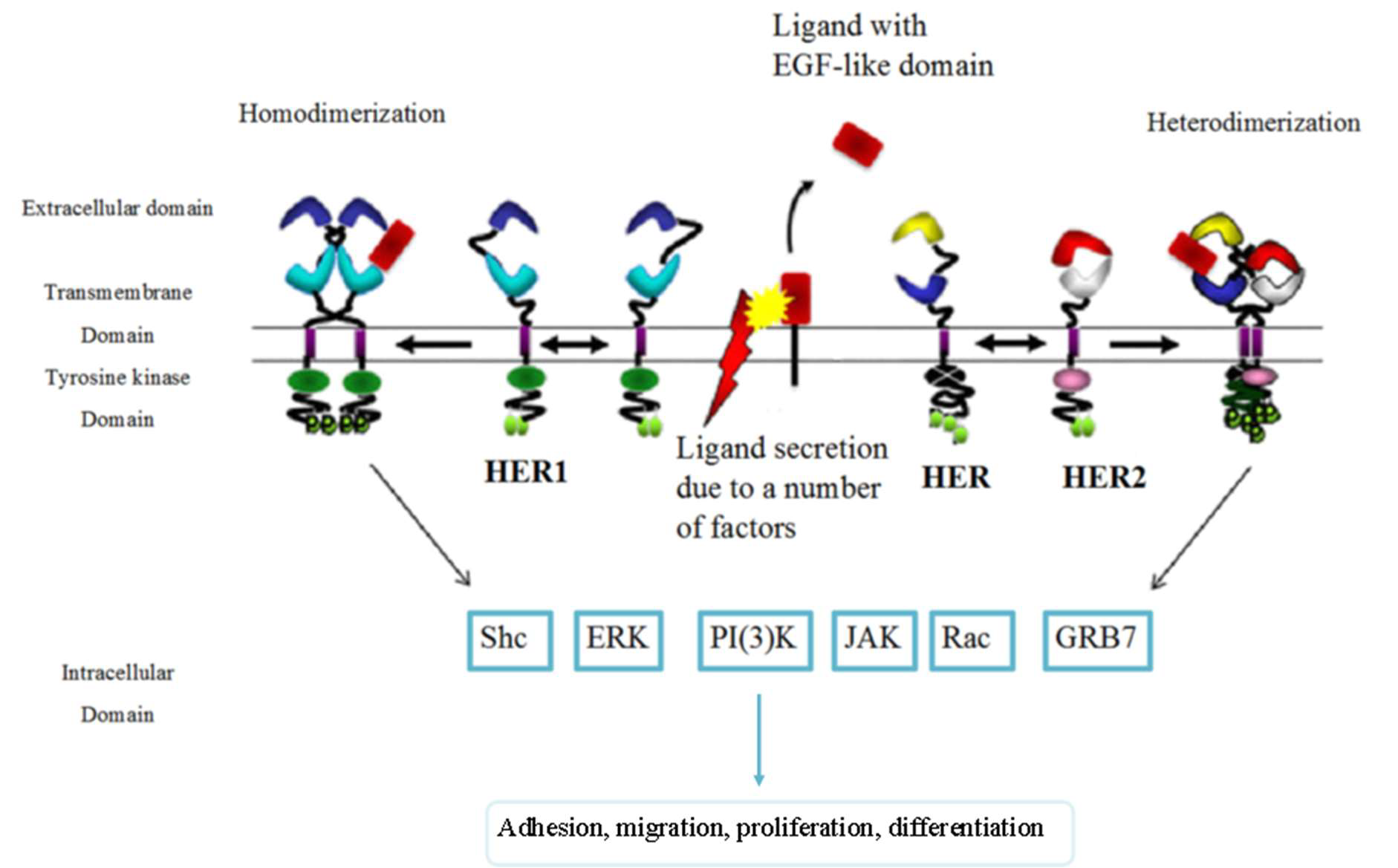
The epidermal growth factor receptor family is frequently over-expressed in human cancers. They are one of the 20 families of human receptor tyrosine kinases (RTK). They are 185 kDa transmembrane glycoproteins that get activated by binding with epidermal growth factors (EGF), EGF-like, or neuregulin ligands. The binding of these proteins form homodimers and heterodimers, which activates their intracellular tyrosine kinase domain leading to the activation of several important oncogenes downstream in the signaling cascade [15].
The epidermal growth factor receptor (EGFR) also known as HER1 and ERBB1 has an important role in starting the signaling that directs epithelial cell behavior (an example of which can be found in Figure 1) and subsequently, tumor cells of epithelial origins [16]. EGFR overexpression often results in a poor prognosis and lower survival rates [17]. It has been established for a while now that EGFR presence is inversely correlated with Estrogen Receptors (ER) [18,19], despite the fact that extensive aspect of its function remains unexplored [20], it has proven to be a useful marker for the few subsets of breast cancer in which it has been fully explored (for example triple negative breast cancer [21] and inflammatory breast cancer [22]). HER2, also known as ERBB2, is unique in that it lacks a ligand; its signaling depends upon heterodimer formation [15].
HER2 is over-expressed or its gene is amplified in approximately 20% of breast cancer patients, and is associated with having a bad prognosis, thus requiring anti-HER2 therapy (trastuzumab or lapatinib) to enhance the odds and length of survival [23].
Mucin 1
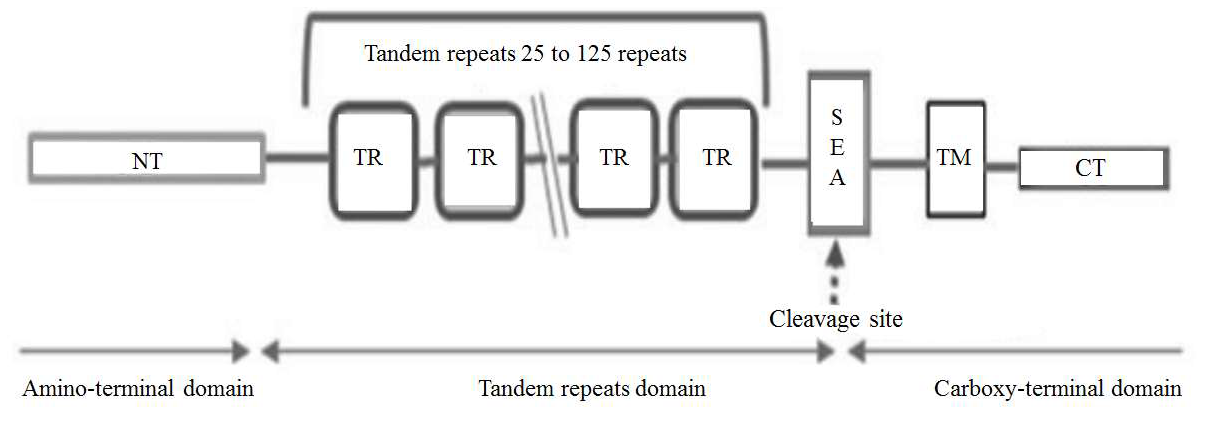
Many different names have been used for Mucin 1 (MUC1) over the last 25 years, the most commonly used name however is the protein’s serum markers for breast cancer CA15.3 and CA27.29. MUC1 is a large cell surface glycoprotein with a molecular weight of around 120 to 225 kDa, this variation is due to the large number of O-linked glycosylation sites on its extracellular domain.
MUC1 go through auto-cleavage resulting in 2 different subunits: the amino terminal subunit (MUC1-N) and the carboxyterminal subunit (MUC1-C or MUC1a). The cleavage occurs at a serine residue near the N-terminal in a GSVVV motif which is located in the extracellular SEA domain. A picture of MUC1 structure can be found in Figure 2 [25].
MUC1’s function is to provide protection against fluctuation of pH and osmolarity, lubrication, and hydration, to the apical surface facing the lumen of ducts by trapping cellular debris on the cell surface. MUC1 can also be found in milk where it acts as a distraction for different types of infective pathogens [26]; its function as a regulatory protein for T-cells is newly discovered and its potential as an immune system regulator is still being discovered [27].
MUC1 is altered in a number of diseases including but not limited to Crohn’s disease, Inflammatory bowel disease, interstitial pneumonitis, and many others. While in cancer, it is overexpressed in several types of cancer from breast and ovarian to prostate, lung and pancreatic cancer. Because of that, it is considered as a universal marker for human cancer.
Because of its universality, many types of serum tests were developed for its detection, chiefly among them are cancer antigen 15-3 (CA 15-3) which is considered a prognostic biomarker for breast cancer worldwide and cancer antigen 27.29 that is used to detect early recurrence of breast and ovarian cancer [26,28]. Its mechanism with breast cancer relates to its ability to aid in immune cell evasion due to MUC1 interactions with siglets or Sialic acid based lectins found on a number of immune cells [29].
In terms of subtypes, there are no evidence found for a link between preoperative CA15-3 levels and different breast cancer subtypes [30].
Tumor protein 53
Tumor Protein 53 (p53) is a remarkable multifunctional protein that plays a role in cell cycle and differentiation, inflammation and immune response, transcription and DNA repair, epigenome, and lastly senescence and autophagy.
In normal cell function, p53 is maintained at low levels, however, being an influential inductor of apoptosis, p53 level increases in response to diverse stress conditions through different posttranslational modifications, causing p53 stabilization and activation leading to apoptosis [31].
In breast cancer, p53 has been in case studies from its prognostic and predictive behaviors [32] toward development of several therapeutic strategies relating to it [33].
In regard to different breast cancer types, p53 is mostly overexpressed in triple negative breast cancer while it is less frequently overexpressed in hormone receptor positive breast cancers [34].
Vascular endothelial growth factor
Vascular endothelial growth factor (VEGF) is a cellular signal protein whose function in the body involves the formation of new blood vessels during embryonic development or after injury or to bypass blockage in a vessel, also it aids in muscle growth following exercise [35].
VEGF’s role in cancer is the promotion of vascular leakage, allowing more blood vessels to reach the tumor than normal cells [36].
In Breast Cancer however, elevated levels of VEGF leads to a worse clinical outcome [37,38]. However, VEGF is a key player in triple negative breast cancer, where it helps the cancer to vascularize and proliferate, as such it is an important biomarker for prognosis and predictive behavior for this particular type of cancer [39,40].
Carcinoembryonic antigen
Carcinoembryonic antigen (CEA) is the first ever cancer biomarker discovered in 1965 [41], since then it has fallen out as a cancer biomarker due to its poor specificity in the case of breast cancer [42]. It also stopped being used in metastatic breast cancer due to the large number of false positives it gives [43]. This is despite the fact that recent meta-analysis have shown particular benefit for CEA as a good prognostic biomarker for poorer disease free and overall survival [44], as well as the possibility of CEA to be a good predictive for metastatic behavior of early breast cancer regardless of subtype [45]. However, recent findings shows that CEA is higher in patients with a history of breast cancer in the family than in history without breast cancer [46].
Conclusion
Salivary protein breast cancer biomarker has benefitted greatly from the advancement of detection techniques. However, much remains to be tested; the biggest issues remain is the lack of large scale research to grant legitimacy for the use of saliva as a medium for detection of breast cancer, and a standardization of saliva collection in order to minimize the matrix effect of the salivary matrix. Despite the lack of data regarding salivary protein biomarker and breast cancer, what we understand of these proteins now is sufficient to form the basis of future research and further development of the techniques for their detection.
Conflicts of Interest
The author reports no conflict of interest.
Acknowledgment
The authors acknowledge the financial support from the UK-Lebanon Techhub as part of their Primispot project, as well as the EU H2020 research and innovation program entitled KardiaTool with grant agreement Nº 768686 and from CAMPUS FRANCE program under grant agreement PHC PROCOPE 40544QH.
References
2. Duffy MJ, Harbeck N, Nap M, Molina R, Nicolini A, Senkus E, Cardoso F. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). European journal of cancer. 2017 Apr 1;75:284-98.
3. Kabel AM. Tumor markers of breast cancer: New prospectives. Journal of Oncological Sciences. 2017 Apr 1;3(1):5-11.
4. Porto-Mascarenhas EC, Assad DX, Chardin H, Gozal D, Canto GD, Acevedo AC, Guerra EN. Salivary biomarkers in the diagnosis of breast cancer: A review. Critical reviews in oncology/hematology. 2017 Feb 1;110:62-73.
5. Greabu M, Battino M, Mohora M, Totan A, Didilescu A, Spinu T, Totan C, Miricescu D, Radulescu R. Saliva- -a diagnostic window to the body, both in health and in disease. Journal of medicine and life. 2009;2(2):124-32.
6. Murthykumar K. Saliva composition and function: a review. International Journal of Pharmaceutical Science and Health Care. 2014;3(4):72-7.
7. Chiu ML, Lawi W, Snyder ST, Wong PK, Liao JC, Gau V. Matrix effects—a challenge toward automation of molecular analysis. JALA: Journal of the Association for Laboratory Automation. 2010 Jun;15(3):233-42.
8. Topkas E, Keith P, Dimeski G, Cooper-White J, Punyadeera C. Evaluation of saliva collection devices for the analysis of proteins. Clinica Chimica Acta. 2012 Jul 11;413(13-14):1066-70.
9. Streckfus C, Bigler L, Tucci M, Thigpen JT. A preliminary study of CA15-3, c-erbB-2, epidermal growth factor receptor, cathepsin-D, and p53 in saliva among women with breast carcinoma. Cancer investigation. 2000 Jan 1;18(2):101-9.
10. Streckfus CF, Arreola D, Edwards C, Bigler L. Salivary protein profiles among HER2/neu-receptorpositive and-negative breast cancer patients: Support for using salivary protein profiles for modeling breast cancer progression. Journal of oncology. 2012;2012.
11. Laidi F, Bouziane A, Lakhdar A, Khabouze S, Rhrab B, Zaoui F. Salivary expression of soluble HER2 in breast cancer patients with positive and negative HER2 status. OncoTargets and therapy. 2014;7:1285.
12. Laidi F, Bouziane A, Errachid A, Zaoui F. Usefulness of salivary and serum auto-antibodies against tumor biomarkers HER2 and MUC1 in breast cancer screening. Asian Pac. J. Cancer Prev. 2016;17(1):335-9.
13. Brooks MN, Wang J, Li Y, Zhang R, Elashoff D, Wong DT. Salivary protein factors are elevated in breast cancer patients. Molecular medicine reports. 2008 May 1;1(3):375-8.
14. Laidi F, Bouziane A, Lakhdar A, Khabouze S, Amrani M, Rhrab B, Zaoui F. Significant correlation between salivary and serum Ca 15-3 in healthy women and breast cancer patients. Asian Pac J Cancer Prev. 2014 Jan 1;15(11):4659-62.
15. Omenn GS, Guan Y, Menon R. A new class of protein cancer biomarker candidates: differentially expressed splice variants of ERBB2 (HER2/neu) and ERBB1 (EGFR) in breast cancer cell lines. Journal of proteomics. 2014 Jul 31;107:103-12.
16. Herbst RS. Review of epidermal growth factor receptor biology. International Journal of Radiation Oncology* Biology* Physics. 2004 Jun 1;59(2):S21-6.
17. Gonzalez-Conchas GA, Rodriguez-Romo L, Hernandez-Barajas D, Gonzalez-Guerrero JF, Rodriguez-Fernandez IA, Verdines-Perez A, Templeton AJ, Ocana A, Seruga B, Tannock IF, Amir E. Epidermal growth factor receptor overexpression and outcomes in early breast cancer: a systematic review and a metaanalysis. Cancer treatment reviews. 2018 Jan 1;62:1-8.
18. Richard J, Sainsbury C, Needham G, Farndon J, Malcolm A, Harris A. Epidermal-growth-factor receptor status as predictor of early recurrence of and death from breast cancer. The Lancet. 1987 Jun 20;329(8547):1398- 402.
19. Nicholson S, Richard J, Sainsbury C, Halcrow P, Kelly P, Angus B, Wright C, Henry J, Farndon JR, Harris AL. Epidermal growth factor receptor (EGFr); results of a 6 year follow-up study in operable breast cancer with emphasis on the node negative subgroup. British journal of cancer. 1991 Jan;63(1):146.
20. Kjaer IM, Bechmann T, Brandslund I, Madsen JS. Prognostic and predictive value of EGFR and EGFRligands in blood of breast cancer patients: a systematic review. Clinical Chemistry and Laboratory Medicine (CCLM). 2018 Apr 25;56(5):688-701.
21. Burness ML, Grushko TA, Olopade OI. Epidermal growth factor receptor in triple-negative and basallike breast cancer: promising clinical target or only a marker?. The Cancer Journal. 2010 Jan 1;16(1):23-32.
22. Wang X, Reyes ME, Zhang D, Funakoshi Y, Trape AP, Gong Y, Kogawa T, Eckhardt BL, Masuda H, Pirman Jr DA, Yang P. EGFR signaling promotes inflammation and cancer stem-like activity in inflammatory breast cancer. Oncotarget. 2017 Sep 15;8(40):67904.
23. Wang T, Zhou J, Zhang S, Bian L, Hu H, Xu C, Hao X, Liu B, Ye Q, Liu Y, Jiang Z. Meaningful interpretation of serum HER2 ECD levels requires clear patient clinical background, and serves several functions in the efficient management of breast cancer patients. Clinica Chimica Acta. 2016 Jul 1;458:23-9.
24. Finigan JH, Downey GP, Kern JA. Human epidermal growth factor receptor signaling in acute lung injury. American journal of respiratory cell and molecular biology. 2012 Oct;47(4):395-404.
25. Jahan R, Kaur S, Macha MA, Batra S. Encyclopedia of Signaling Molecules. Encycl Signal Mol. 2016.
26. Apostolopoulos V, Stojanovska L, Gargosky SE.MUC1 (CD227): a multi-tasked molecule. Cellular and molecular life sciences. 2015 Dec 1;72(23):4475-500.
27. Agrawal B, Gupta N, Konowalchuk JD. MUC1 Mucin: A Putative Regulatory (Checkpoint) Molecule of T Cells. Frontiers in immunology. 2018;9.
28. Jing X, Liang H, Hao C, Yang X, Cui X. Overexpression of MUC1 predicts poor prognosis in patients with breast cancer. Oncology reports. 2019 Feb 1;41(2):801-10.
29. Kuol N, Stojanovska L, Nurgali K, Apostolopoulos V.The mechanisms tumor cells utilize to evade the host’s immune system. Maturitas. 2017 Nov 1;105:8-15.
30. Araz M, Beypinar I, Kazan S, Inci F, Celiker M, Uysal M. Are preoperative serum CA15-3 levels different in breast cancer subgroups?. Current problems in cancer. 2019 Apr 1;43(2):115-22.
31. Issaeva N, Mediating J. p53 Signaling in Cancers. Cancers. 2019 Mar; 11(3): 332.
32. Gasco M, Shami S, Crook T. The p53 pathway in breast cancer. Breast cancer research. 2002 Apr 1;4(2):70.
33. Lacroix M, Toillon RA, Leclercq G. p53 and breast cancer, an update. Endocrine-related cancer. 2006 Jun 1;13(2):293-325.
34. Qamar S, Khokhar MA, Farooq S, Ashraf S, Humayon WA, Rehman A. Association of p53 Overexpression with Hormone Receptor Status and Triple Negative Breast Carcinoma. Journal of the College of Physicians and Surgeons Pakistan. 2019 Feb 1;29(2):164-7.
35. Laddha AP, Kulkarni YA. VEGF and FGF-2: Promising targets for the treatment of respiratory disorders. Respiratory medicine. 2019 Aug 8.
36. Stuttfeld E, Ballmer-Hofer K. Structure and function of VEGF receptors. IUBMB life. 2009 Sep;61(9):915-22.
37. Banys-Paluchowski M, Witzel I, Riethdorf S, Pantel K, Rack B, Janni W, Fasching PA, Aktas B, Kasimir-Bauer S, Hartkopf A, Solomayer EF. The clinical relevance of serum vascular endothelial growth factor (VEGF) in correlation to circulating tumor cells and other serum biomarkers in patients with metastatic breast cancer. Breast cancer research and treatment. 2018 Nov 1;172(1):93-104.
38. Baker L, Hall L, Wilson D, Bhaskar P. Plasma levels of mammaglobin-A, VEGF and PlGF in human breast cancer pathology and survival. European Journal of Surgical Oncology. 2019 Feb 1;45(2):e40.
39. Wang RX, Chen S, Huang L, Zhou Y, Shao ZM. Monitoring Serum VEGF in Neoadjuvant Chemotherapy for Patients with Triple-Negative Breast Cancer: A New Strategy for Early Prediction of Treatment Response and Patient Survival. The oncologist. 2019 Jun 1;24(6):753-61.
40. Hwang SY, Park S, Kwon Y. Recent therapeutic trends and promising targets in triple negative breast cancer. Pharmacology & therapeutics. 2019 Jul 1;199:30-57.
41. Mitchell JS, Lowe TE. Matrix effects on an antigen immobilized format for competitive enzyme immunoassay of salivary testosterone. Journal of immunological methods. 2009 Sep 30;349(1-2):61-6.
42. Robertson JF, Pearson D, Price MR, Selby C, Pearson J, Blamey RW, Howell A. Prospective assessment of the role of five tumour markers in breast cancer. Cancer Immunology, Immunotherapy. 1991 Nov 1;33(6):403- 10.
43. Gam LH. Breast cancer and protein biomarkers. World journal of experimental medicine. 2012 Oct 20;2(5):86.
44. Li X, Dai D, Chen B, Tang H, Xie X, Wei W.Clinicopathological and Prognostic Significance of Cancer Antigen 15-3 and Carcinoembryonic Antigen in Breast Cancer: A Meta-Analysis including 12,993 Patients. Disease markers. 2018;2018.
45. Imamura M, Morimoto T, Nomura T, Michishita S, Nishimukai A, Higuchi T, Fujimoto Y, Miyagawa Y, Kira A, Murase K, Araki K. Independent prognostic impact of preoperative serum carcinoembryonic antigen and cancer antigen 15-3 levels for early breast cancer subtypes. World journal of surgical oncology. 2018 Dec;16(1):26.
46. Usoro AJ, Udoh AE, Usoro CA, Etuk EB, Obot AS. Evaluation of Serum Levels of Mammaglobin, Carcinoembryonic Antigen, Prolactin, Estradiol and Free Prostate Specific Antigen As Biomarkers of Breast Cancer.