Abstract
The ultrasensitive ELISA method developed by Watanabe and Ito combines sandwich ELISA and thio-NAD cycling to enable the quantitation of trace amounts of proteins. The ultra-traceability provided by this method makes it possible to quantify extremely small amounts of proteins in small extracellular vesicles called exosomes as well as in urine. As a result, rather than relying on ultrasensitive detection of nucleic acids, the functions of the proteins directly involved in cancer can be clarified. In the present commentary, we discuss the contribution of the ultrasensitive ELISA to the advancement of cancer research.
Keywords
Adiponectin, Cancer, Exosome, GPR78, Ultrasensitive ELISA, thio-NAD cycling, Urine
Commentary
Much of cancer research is centered on the analysis of nucleic acids [1]. DNA testing is considered to be the gold standard in cancer testing [2]. This is because even very small amounts (i.e., only a few copies) of nucleic acids can be amplified by PCR [3]. Is this, however, really the best option for cancer research? In many cases, it is not nucleic acids but proteins that act directly on the cancer cells and tissues. In this case, it is important to accurately determine and quantify the proteins that are specifically produced and released by cancer cells, but this is quite difficult. The reason for the difficulty is that many of the proteins released by cancer cells are produced in extremely trace amounts, often undetectable by conventional protein detection methods. Therefore, a measurement method for detecting extremely small amounts of protein is needed.
Ultrasensitive ELISA for Detecting Trace Amounts of Proteins
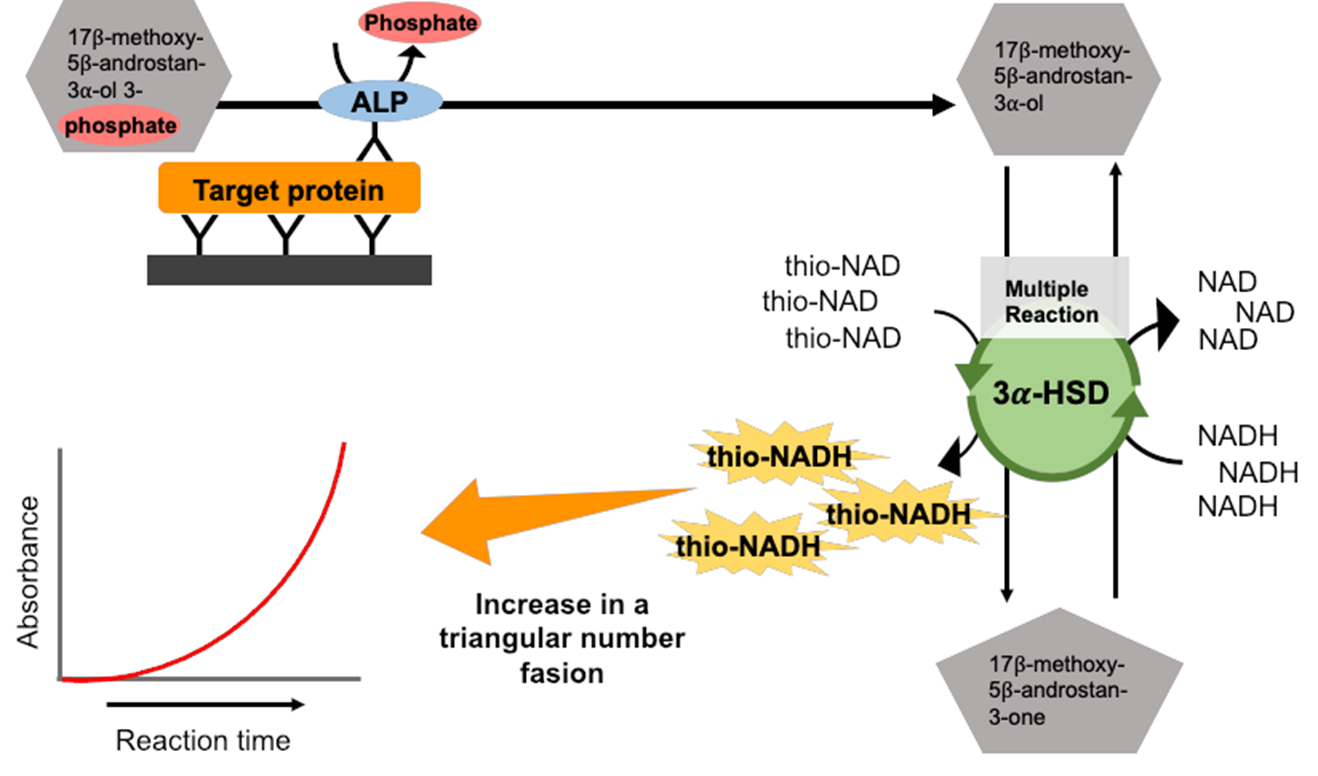
To achieve ultrasensitive, quantitative detection of proteins, Watabe and Ito attempted to realize an ultrasensitive ELISA by combining a sandwich ELISA with thio-nicotinamide adenine dinucleotide (thio-NAD) cycling [4]. The major challenge for protein quantification is that, unlike nucleic acids, proteins cannot be amplified using a method such as PCR. We hypothesized that we could amplify the detection signal of specific proteins and designed a method for amplifying the signals produced by the antigen-antibody reaction in an ELISA.
The most suitable quantification technique for a trace amount of protein is a sandwich ELISA [5]. A standard sandwich ELISA produces a color change in the substrate, resulting in a detectable signal. This signal increases in a linear fashion over time, and thus the sensitivity is limited, which is a weak point of conventional ELISAs. We considered that a substrate that is hydrolyzed by alkaline phosphate (ALP) could be amplified, thereby providing the necessary signal amplification.
We identified another assay for detecting a trace amount of molecules that utilizes an enzyme cycling method to amplify the signal [6]. Lowry wrote in 1980: “Enzymatic cycling provides a methodology for virtually unlimited amplification of analytical sensitivity. The most widely applicable cycling systems are those for NAD and NADP, since these can be used to increase the sensitivity of methods for a host of other substances” [7]. We thus considered that the combination of a sandwich ELISA with enzyme cycling could provide an ultrasensitive ELISA [8].
We used single-enzyme cycling with 3α-hydroxysteroid dehydrogenase (3α-HSD) [9]. The 3α-HSD catalyzes the substrate cycling between 3α-hydroxysteroid and its corresponding 3-ketosteroid in the presence of an excess amount of NADH and thio-NAD as cofactors of 3α-HSD. When androsterone 3-phosphate is used as the first substrate (Figure 1), androsterone corresponding to 3α-hydroxysteroid is produced by the hydrolysis of androsterone 3-phosphate via ALP, and androstane 3,17-dione corresponding to 3-ketosteroid is produced via 3α-HSD with NADH as a cofactor. Then, 3-ketosteroid (androstane 3,17-dione) returns to 3α-hydroxysteroid (androsterone) via 3α-HSD with thio-NAD as a cofactor. This series of reactions is called ‘thio-NAD cycling’ [10]. In this reaction, thio-NAD is reduced to thio-NADH, and the accumulated thio-NADH is measured at an absorbance of 405 nm without any interference by the absorbance of NAD, NADH, and thio-NAD.
We combined these 2 assays, sandwich ELISA and thio-NAD cycling. This combination results in the phenomenon that the number of thio-NAD molecules consumed in each cycling step increases linearly over time, whereas thio-NADH accumulates in a triangle number fashion:
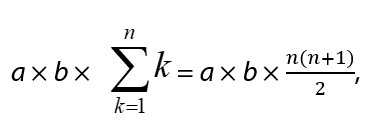
wherein a is the turnover ratio of ALP per minute, b is the cycling ratio of 3α-HSD per minute, and n is the measurement time in minutes. Currently, ALP bound to the secondary antibody in a sandwich ELISA can be detected at the zeptomole (i.e., 10−21 moles) level [11].
What Does the Term ‘Ultrasensitive’ Mean?
As described above, we attempted to develop an ultrasensitive ELISA consisting of a sandwich ELISA and thio-NAD cycling to overcome the detection limits of conventional ELISAs. The term ‘ultrasensitive’ is not well defined, so what do we mean by ‘ultrasensitive’ in this case? One reason that the definition of ultrasensitive is unclear is that it is compared to the detection limits of PCR. PCR can amplify nucleic acids, resulting in the detection of a small number of DNA or RNA copies. For example, the detection limit of real-time PCR is a few copies/assay for various nucleic acids [3]. The limit of detection cannot be compared between real-time PCR and protein measurement systems in which the proteins are not amplified. Thus, it is inaccurate and misleading to state that the detection limit of protein measurement systems is inferior to that of PCR and that the word ‘ultrasensitive’ should not be used for protein measurements.
A second reason for the misinterpretation of the term ‘ultrasensitive’ originates from a comparison of the limit of detection among various other methods used for protein measurement. As described above, many techniques can be used to quantify target proteins, including ELISA, liquid chromatography-mass spectrometry, and lab-on-a-chip methods. Whereas the method with the lowest detection limit might be considered an ‘ultrasensitive’ method and superior to others, “ultrasensitive is not appropriate in this case, because it is useless to compare methods solely on the basis of detection limits without also considering the need for a special apparatus or a special room, as well as the cost. That is, the term ‘ultrasensitive’ does not imply a specific value. So, then, what exactly is ‘ultrasensitive’?
Professor Gooding from the University of New South Wales, Australia, addressed this question in detail [13]. The term ‘ultrasensitive’ is used differently between chemists and biologists. To define the term in an interdisciplinary manner, Gooding defined an ultrasensitive bioanalytical sensor as “a biosensor with sufficient sensitivity and sufficiently low background to allow subpicomolar detection limits”. He also explained that “this is a compromise between the chemical and biological meaning of sensitivity that acknowledges the link between the chemical meaning of a detection limit, and sensitivity as it relates to the background signal”. A limit of detection of 10-13 moles/L or smaller can be referred to as ‘ultrasensitive’ detection. If we use a 96-well microplate for ELISA, 1 assay volume is generally equal to 100 μL. Thus, 10-13 moles/L is equivalent to 10-17 moles/assay, which might be challenging to achieve. Our ELISA method, however, can detect proteins at zeptomolar detection limits, i.e., 10-21 moles/assay [11], and thus, we believe that our system can be classified as an ‘ultrasensitive’ ELISA.
Contribution of This Ultrasensitive ELISA to Cancer Research
Proteins in exosomes may play an important role in the communication between cancer cells (or between cancer cells and normal cells) in the tumor microenvironment [14]. Measuring trace amounts of proteins in exosomes is difficult, however, and thus the cancer stemness-promoting mechanisms of exosomal proteins have not been elucidated. Our team attempted to quantify trace amounts of 78-kDa glucose-regulated protein (GRP78), which is involved in cancer progression, in exosomes released from cultured gastric cancer cells using the ultrasensitive ELISA [14]. We also evaluated the cancer stemness-promoting effects by applying high-GRP78-containing exosomes to cultured gastric cancer cells. The ultrasensitive ELISA enabled the detection of GRP78 at a limit of detection of 1.99 × 10-19 moles/assay (0.16 pg/mL). The stemness of cultured cancer cells incubated with high-GRP78-containing exosomes obtained from GRP78-overexpressed cells was increased on the basis of both an MTT assay and a wound healing assay. Our results demonstrated that the ultrasensitive ELISA has a strong potential for measuring trace amounts of exosomal proteins. Further, exosomes with a high concentration of GRP78 promoted the cancer stemness of the surrounding cells.
As described above, exosomes containing GRP78 are involved in cancer malignancy. Although GRP78 is thought to promote the tumor microenvironment, leading to angiogenesis, no direct evidence for this role has been reported, mainly because of the difficulties in accurately measuring the GRP78 concentration in exosomes. Thus, we quantified the GRP78 concentrations in exosomes collected from gastric cancer AGS cells with GRP78 overexpression, GRP78 knockdown, or GRP78 mock [15]. These 3 types of exosomes were then incubated with vascular endothelial cells to examine their effects on endothelial cell angiogenesis. Based on the results of a tube formation assay, GRP78-overexpressed exosomes accelerated angiogenesis compared with GRP78-knockdown or GRP78-mock exosomes. To investigate the mechanisms underlying this effect, we examined the Ser473 phosphorylation state ratio of AKT, which is involved in the angiogenesis process, and found that AKT phosphorylation was increased by applying exosomes overexpressing GRP78 to the endothelial cells. An MTT assay showed that treatment with exosomes overexpressing GRP78 increased the endothelial cell proliferation rate, and a wound healing assay showed that this treatment increased the endothelial cell migration capacity. Therefore, our ultrasensitive ELISA clarified that GRP78-containing exosomes promoted the tumor microenvironment and induced angiogenesis.
Furthermore, the ultrasensitive ELISA that we developed successfully measured trace amounts of proteins, even after separating the lumen and membrane fractions of the exosomes [12]. To develop an adequate and easy-to-use lumen and membrane fraction separation method, we applied a commercially available kit originally developed for cells to exosomes and examined the validity of the results compared with those obtained using a conventional, complicated Na2CO3 method. GRP78 was successfully quantified in both the lumen and membrane fractions of exosomes obtained from cultured cancer cells. These results will facilitate studies to broaden our understanding of the tumor microenvironment.
In general, proteins present in the blood may also be detected in the urine. The protein concentrations in the urine are much lower than those in the blood, however, making it difficult in many cases to apply ELISAs optimized for blood proteins to urine proteins. Adiponectin is an adipocyte-derived vasoactive peptide with anti-inflammatory and insulin-sensitizing peptides that is increased in individuals with obesity and diabetes mellitus, but it is not a biomarker for cancer [16]. Serum adiponectin is believed to enhance insulin sensitivity, and individuals with obesity and type 2 diabetes mellitus often have low serum adiponectin levels. In contrast to the serum levels, our measurements of urinary adiponectin with the ultrasensitive ELISA showed that the urinary adiponectin levels were higher in those with diabetes mellitus than in normal subjects [16]. That is, we validated the use of urinary adiponectin as a marker of the progression of diabetic nephropathy. The sensitivity of commercially available ELISA kits, however, is insufficient for measuring the low levels of urinary adiponectin in normal subjects. Using the ultrasensitive ELISA, we could sufficiently detect the low urinary adiponectin levels in normal subjects, providing a noninvasive test for diabetic nephropathy.
One of the authors (D.M.) is currently working on detecting human papillomavirus (HPV)-related proteins in the urine of cervical cancer patients. The preliminary results suggest that HPV-related proteins can be detected in patient urine using the ultrasensitive ELISA and may be applied as a new noninvasive test for cervical cancer.
Conclusions
Many researchers and physicians are well aware of the importance of studying proteins rather than just nucleic acids in cancer research. Conducting protein research, however, is very difficult. We thus proposed an ultrasensitive ELISA and were successful in detecting trace amounts of protein (<10-17 moles/assay) with an easy-to-use colorimetric method, which had not been achieved previously. In fact, this ultrasensitive detection allows us to quantify very small amounts of proteins contained in very small amounts of exosomes, which means that only a small volume of exosome samples is needed. Furthermore, the ability to detect extremely small amounts of proteins in urine has opened the way for noninvasive testing that does not require blood sampling. Thus, our ultrasensitive ELISA will make a large contribution to cancer research.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Funding Statement
This research was funded by SPRING from JST (JPMJSP2128) to K.I., a Grant-in-Aid for Young Scientists (Early Bird) from the Waseda Research Institute for Science and Engineering, Waseda University, to K.I., and the START Program from JST (JPMJST2053) to E.I. The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
References
2. https://www.who.int/europe/news/item/11-09-2021-who-recommends-dna-testing-as-a-first-choice-screening-method-for-cervical-cancer-prevention
3. Wagatsuma A, Sadamoto H, Kitahashi T, Lukowiak K, Urano A, Ito E. Determination of the exact copy numbers of particular mRNAs in a single cell by quantitative real-time RT-PCR. J Exp Biol. 2005 Jun;208(Pt 12):2389-2398.
4. Watabe S, Kodama H, Kaneda M, Morikawa M, Nakaishi K, Yoshimura T, et al. Ultrasensitive enzyme-linked immunosorbent assay (ELISA) of proteins by combination with the thio-NAD cycling method. Biophysics. 2014 Sep 5;10:49-54.
5. Tsurusawa N, Chang J, Namba M, Makioka D, Yamura S, Iha K, et al. Modified ELISA for ultrasensitive diagnosis. J Clin Med. 2021 Nov 7;10(21):5197.
6. Kato T, Berger SJ, Carter JA, Lowry OH. An enzymatic cycling method for nicotinamide-adenine dinucleotide with malic and alcohol dehydrogenases. Anal Biochem. 1973 May;53(1):86-97.
7. Lowry OH. Amplification by enzymatic cycling. Mol Cell Biochem. 1980 Nov 20;32(3):135-146.
8. Ito E, Iha K, Yoshimura T, Nakaishi K, Watabe S. Early diagnosis with ultrasensitive ELISA. Adv Clin Chem. 2021;101:121-133.
9. Kyosei Y, Namba M, Makioka D, Kokubun A, Watabe S, Yoshimura T, et al. Ultrasensitive detection of SARS-CoV-2 spike proteins using the thio-NAD cycling reaction: A preliminary study before clinical trials. Microorganisms. 2021 Oct 25;9(11):2214.
10. Iha K, Inada M, Kawada N, Nakaishi K, Watabe S, Tan YH, et al. Ultrasensitive ELISA developed for diagnosis. Diagnostics. 2019 Jul 18;9(3):78.
11. Iha K, Kyosei Y, Namba M, Makioka D, Yamura S, Watabe S, et al. Zeptomole detection of an enzyme by a simple colorimetric method. Anal Sci. 2021 Oct 10;37(10):1469-1472.
12. Iha K, Tsurusawa N, Tsai HY, Lin MW, Sonoda H, Watabe S, et al. Ultrasensitive ELISA detection of proteins in separated lumen and membrane fractions of cancer cell exosomes. Anal Biochem. 2022 Oct 1;654:114831.
13. Gooding JJ. What does ultrasensitive really mean? ACS Sens. 2019 Mar 22;4(3):528.
14. Tsurusawa N, Iha K, Sato A, Tsai HY, Sonoda H, Watabe S, et al. Ultrasensitive detection of GRP78 in exosomes and observation of migration and proliferation of cancer cells by application of GRP78-containing exosomes. Cancers. 2022 Aug 11;14(16):3887.
15. Iha K, Sato A, Tsai HY, Sonoda H, Watabe S, Yoshimura T, et al. Gastric cancer cell-derived exosomal GRP78 enhances angiogenesis upon stimulation of vascular endothelial cells. Curr Issues Mol Biol. 2022 Dec 6;44(12):6145-6157.
16. Yamakado S, Cho H, Inada M, Morikawa M, Jiang YH, Saito K, et al. Urinary adiponectin as a new diagnostic index for chronic kidney disease due to diabetic nephropathy. BMJ Open Diabetes Res Care. 2019 May 30;7(1):e000661.