Abstract
Background: Coxiella burnetii is the causative agent of Q fever, a human disease that can be acquired from livestock. Diseases caused by this organism have caused great losses in livestock and human health. No vaccine is approved for use in the United States, and formalin-inactivated whole-cell vaccines pose a significant manufacturing risk for biocontainment. A subunit vaccine using recombinant peptides from C. burnetii would be safer and less resource-intensive to produce. This study used reverse vaccinology to expand our prediction sets of T-cell epitopes for the major histocompatibility complex (MHC) Class I and II alleles of cattle, sheep, and goats. Thereafter, the present results were compared with those from our previous prediction sets for mice and humans.
Results: Small ruminant breed representation for the United States was ensured by querying whole genome sequences on the National Center for Biotechnology Information database. Consequently, twenty-two sheep MHC Class I, seventeen goat MHC Class I, and one goat MHC Class II alleles were added to the analyses, resulting in a total of fifty-six sheep MHC Class I, eighteen goat MHC Class II, and twenty-seven goat MHC Class II alleles. Predicted interactions of C. burnetii proteome-derived peptides with each MHC allele were categorized as strong, weak, or non-binding based on bioinformatic scores. Interspecies comparisons resulted in 256 peptides of interest for MHC Class II presentation and 766 peptides of interest for MHC Class I presentation. Of these, 51 peptides were predicted to bind with both classes of MHC alleles, of which 33 were newly identified.
Conclusion: The high scoring T-cell epitope predictions identified in this study provide grounds for prioritizing subunit candidates to further develop a safe and perhaps broadly effective C. burnetii vaccine.
Keywords
Coxiella burnetii, T-cell epitope, Small ruminant, Bioinformatics, Reverse vaccinology
Introduction
Subunit vaccines contain specific peptides known to confer protection, alleviate disease symptoms, or prevent spread [1-4]. This approach to vaccine development requires knowledge about which pathogen peptides elicit such responses in the vaccinated host. Without this foundational knowledge, vaccine development proceeds fastest when the pathogen of interest has a small proteome or an antibody-mediated protective response. An example of this is influenza, where antibodies against hemagglutinin or neuraminidase reduce disease symptoms, spreading, and time course [4,5]. On the other hand, effective immune responses to some pathogens require cell-mediated immunity, and direct peptide investigation for many pathogens is hampered by the sheer size of their proteomes [6-10]. One such pathogen is Coxiella burnetii, a bacterium that produces thousands of proteins and for which effective defenses against infection in humans and animals requires cell-mediated immune responses [11,12].
In humans, exposure to C. burnetii can result in several outcomes, including asymptomatic transient infection, acute illness, or persistent infection [11,13]. Estimation of case outcomes would suggest that 40% of exposed humans respond with acute flu-like illness while the rest of the exposed population are asymptomatic [11,13,14]. Chronic Q fever and persistent Q fever are terms used to designate long-term symptomatic infections in humans [11,13], which commonly present as either endocarditis, osteoarticular infection, hepatitis, or vascular infection [11,13,14]. Infections in animals may also remain asymptomatic but the most common clinical outcome is reproductive disease [12,15-17]. Importantly, asymptomatic infections in domestic ruminants can still result in high bacterial shedding in their birth products [15,16,18]. Thus, C. burnetii is of concern to both animal and human health, and the risk of exposure is most critical in the vicinity of periparturient domestic ruminants.
Coxiella burnetii is a Gram-negative bacterium that readily persists within the environment in a spore-like form and is considered endemic in many regions of the world [19,20]. The respiratory tract is the most common route of exposure for animals and humans alike, and by which the minimum infective dose is very low [18,21]. In light of these facts and the risk of severe clinical outcomes, C. burnetii is classified as a Biosafety Level 3 (BSL3) organism [21]. Thus, producing a formalin-inactivated whole-cell C. burnetii vaccine requires expensive BSL3 laboratories and procedures and has not been approved in the United States. Without an effective vaccine, endemic areas like the United States remain at high risk for outbreaks [22-24]. An epidemic outbreak of Coxiella burnetii occurred in the Netherlands between 2007 and 2010, which cost the country millions of dollars to treat human cases and implement emergency animal intervention strategies [25]. It is therefore prudent to continue efforts toward developing an effective C. burnetii vaccine that can be efficiently and safely produced.
Reverse vaccinology is a technique that has gained interest in recent years [4,9,26]. This technique relies on bioinformatic tools to assess the ability of pathogen proteins to activate the host’s immune system. Activation of T-cells relies on only the primary structure (i.e., linear sequence) of a pathogen protein, whereas B-cell activation can require recognition of higher-order tertiary protein structure. Further, T-cell activation relies on the interaction between a given pathogen peptide and the major histocompatibility complex (MHC), a complex of host proteins that establish an adaptive immune response by presenting peptides of pathogen proteins to T-cells. Peptides that interact with MHCs are designated T-cell epitopes. There are two basic types of T-cells, wherein each responds to a different allelic class of MHC molecule. MHC Class I molecules present bound peptides to ‘cytotoxic’ (CD8+) T-cells whereas MHC Class II molecules present bound peptides to ‘helper’ (CD4+) T-cells. By utilizing MHC eluted ligands and known amino acid interaction preferences between peptides and MHC alleles, modern bioinformatic tools mine the exceptionally large sequence space of pathogens with large proteomes for peptides that are most likely to activate T-cell mediated host immune responses.
Bioinformatic tools have been recently expanded to include MHC alleles of cattle [27,28]. Our group previously applied these tools to predict C. burnetii peptides that bind the known sequences of MHC Class I in cattle, as well as MHC Class I and Class II alleles within the mouse and human species [29]. A bovine MHC Class II prediction tool, NetBoLAIIpan, has subsequently become available [27]. In the present analysis, NetBoLAIIpan is employed to expand predictions for cattle, sheep, and goat T-cell epitopes within the C. burnetii proteome. In addition, NetMHCpan was utilized to predict MHC Class I T-cell epitopes for sheep and goat alleles.
Methods
Random peptide generation
Protein sequences were randomly generated using the Expasy RandSeq tool [30], where sequences were produced using the average amino acid composition present on Swiss-Prot. Generated proteins were separated into 13-mer, 14-mer, 15-mer, 16-mer, or 17-mer peptides to be scrambled. Mimotopes Scrambled tool was exploited to scramble supplied amino acids one thousand times [31]. Forty-thousand peptides were produced for each peptide length, making a total of 200,000 randomly generated peptides to train NetBoLAIIpan 1.0 for small ruminant allele evaluation (Personal Communication).
MHC Class I epitopes are usually shorter than MHC Class II epitopes [27,32-34], so downsizing of random epitopes generated for MHC Class II was used to supply MHC Class I epitopes. Generation of random peptides consisting of 8-mers, 9-mers, 10-mers, and 11-mers was accomplished using the longer random peptide sequences. Specifically, peptides from 13-mer, 14-mer, 15-mer, and 16-mer files had 5 amino acids removed from the C-terminal end to generate 160,000 random peptides used to train NetMHCpan 4.1.
Identification of MHC Class I and MHC Class II alleles from sheep and goat whole genome sequences
NCBI was assessed for whole genome sequences pertaining to domestic sheep and goat species. Present whole genome sequences (WGS) were only included in the analysis if the assembly had reached chromosome level, 25X coverage, and was in a breed of interest (Supplemental Table 1). Supplemental Table 1 contains the species, breed, chromosome accession number, and MHC classes evaluated within present WGSs. Isolated WGSs were evaluated for MHC alleles within chromosome 20 for domestic sheep species and within chromosome 23 for domestic goat species, as these chromosomes are known to house the MHC haplotypes [35]. Tblastn was employed to assess ten million base pairs of genome sequence at a time for the presence of MHC alleles. The protein query used was Ovar-N*01:01 (CAI43967.2) for MHC Class I within domestic sheep, ABQ14768.1 for MHC Class I within domestic goat, and Cahi-DRB1*01:01 (SPC50560.1) for MHC Class II within domestic goat [36-39]. Outputs were scrutinized for genomic regions which might house MHC alleles using the graphics feature, where an overall hit that had an identity above 50% was scrutinized for a grouping of hits within the graphics feature. If one of the hits within a group had an identity above 78%, then the million base pair region was marked for further analysis.
Following notation of these regions, Genscan was used to translate DNA [40]. Predicted proteins were queried using the Basic Local Alignment Search Tool for protein (BLASTp) and the previously mentioned MHC Class I or MHC Class II alleles. Returned hits were scrutinized for percent identity and percent coverage indicative of true MHC alleles. To determine cut-off values, known Immuno Polymorphism database (IPD)-MHC database alleles were queried using BLASTp. This analysis determined that true sheep MHC Class I alleles tend to have a percent identity above 78% and a percent coverage above 80%. Concurrently, goat MHC Class II alleles were found to maintain a percent identity above 75% and a percent coverage of 30%. However, when assessing translated proteins comparatively against Cahi-DRB1*01:01, a percent coverage of 80% was employed due to most of the annotated goat MHC Class II alleles being partially sequenced. With only one prior MHC Class I allele present for goat species, a percent identity of 75% and a percent coverage of 80% was employed to follow what was determined for goat MHC Class II alleles.
Following MHC allele identification, pairwise distance analysis of allele proteins from the IPD-MHC database, prior publications, and WGS inquiry was conducted [36-38,40-50]. Pairwise distance used MEGA X software version 10.1.8, where exons 2 and 3 were aligned for MHC Class I alleles and exon 2 was aligned for MHC Class II alleles [51]. Resultant pairwise distance charts are present in Supplemental Files 1 (MHC Class II) and 2 (MHC Class I). A pairwise distance of 0.0 indicated that tested alleles maintained 100% identity. Alleles that were not previously described but identified during whole genome scrutiny have their genomic locus listed as LP within Supplemental Tables 2-4; as well as in Supplemental File 3.
Selection of sheep MHC Class II alleles
Due to the plethora of MHC Class II alleles (130 Ovar-DRB1) present on IPD-MHC and having to train each allele within the NetBoLAIIpan 1.0 program, only a subset was chosen for analysis. Sheep MHC Class II alleles were selected based off breed association, frequency within populations, and phylogenetic distance present within published articles [42-44,46-50,52]. A list of the collected alleles and associated breeds of interest are present in Supplemental Tables 2 and 3.
NetBoLAIIpan version 1.0
Recent release of NetBoLAIIpan 1.0 allows for cattle MHC Class II alleles to be tested for interaction with 15-mer peptides [27]. Present on this database and program are 299 cattle MHC Class II alleles (Supplemental Table 2), which align with the DRB3 locus within the cattle genome. Run proteins were previously described in Piel et. al [29] and were used here to assess cattle MHC Class II allele interaction with the conserved Coxiella burnetii proteome.
To more accurately assess small ruminant MHC Class II alleles for interaction with C. burnetii peptides, the NetBoLAIIpan 1.0 program needed to be trained for each allele of interest by running the 200,000 randomly generated peptides per allele. The DRA gene is typically invariant or nearly so [53,54]. Importantly, the DRA allele used within NetBoLAIIpan 1.0 cannot be altered; therefore, each of the small ruminant MHCII alleles was run using the cattle MHC Class II-DRA and a small ruminant MHC Class II-DRB. Differences between ruminant MHC Class II DRA alleles were defined by protein alignment using ClustalW within the MEGA X software, employed gap and extension penalties were not altered from program defaults. Percent identity was calculated by using the base parameters on BLASTp. Following training, the small ruminant MHC Class II DRB alleles listed in Supplemental Table 3 were tested for T-cell epitopes using the conserved C. burnetii proteome.
Following acquisition of NetBoLAIIpan 1.0 output files for all ruminant species, each file was loaded into the Sequel Server Management Studio (SSMS) v18.6. For cattle, tested MHC Class II alleles had percent rank values calculated by NetBoLAIIpan 1.0. Percent rank values were used to determine if a tested peptide bound strongly (x < 2) or weakly (2 ≤ x < 10) to a given allele. This follows the scoring previously used [29] to define C. burnetii T-cell epitopes within the human and mouse species. The percent rank for small ruminant species was not calculated by NetBoLAIIpan 1.0; instead, the random peptide scores were employed per allele to determine the score associated with the top 2% and top 10% ranked hits. Thereafter, these score denominators were used to define weak and strong binders as done for the cattle species.
NetMHCpan version 4.1
Prior analysis has occurred for the 105 Bovine Leukocyte Antigen (BoLA) MHC Class I alleles present on NetMHCpan 4.1 [29]. Expansion of this dataset to include small ruminants MHC Class I alleles required running 160,000 randomly generated peptides for each allele of interest. Thereafter, each MHC Class I allele defined for sheep and goat species was analyzed against the conserved C. burnetii proteome. Similar to NetBoLAIIpan analysis, the output data files resulting from random peptide analysis were loaded onto a SSMS database. However, for MHC Class I analysis, the scores associated with the top 0.5% and 2% random peptides were defined, where a strong interaction had a percent rank < 0.5 and a weak interaction had a percent rank ≥ 0.5 and < 2.
High scoring peptides from NetBoLAIIpan 1.0 and NetMHCpan 4.1
Within the SSMS databases, percent ranks rendered each peptide:allele interaction as strong, weak, or non-existent. Thereafter, each peptide tested had the number of interactions between every allele summed. Within these summations, peptides which interacted with 45% of the tested alleles strongly or interacted with 90% of the alleles tested were isolated as high scoring peptides for MHC Class II alleles. Alternatively, MHC Class I high scoring peptides were identified as those which interacted with 60% of the alleles tested or were suggested to strongly interact with 45% of the alleles tested. Once the lists of high scoring peptides were calculated, they were scrutinized for peptides which overlapped by 50%. If given peptides overlapped, then the overlapping peptide(s) were removed. Removal of overlapping peptides prioritized cattle strong binding scores due to the bioinformatic program being trained on cattle elution data [27]. If the strong binding scores were the same, then the total number of alleles bound was scrutinized. This resolved most overlapping situations, but the few that required further discrepancy relied on peptide:allele scores from the ovine species.
Results
T-cell epitopes predicted for MHC Class II alleles of cattle
NetBoLAIIpan predicts pathogen peptides that interact with known cattle MHC Class II alleles, designating interacting peptides as potential T-cell epitopes [27]. There are multiple MHC Class II genes within chromosome 23 of cattle, but the DRB3 locus represents the most polymorphic and best studied [45,55-66]. Therefore, the NetBoLAIIpan program was initiated with DR alleles but not DQ or DY alleles. At the time of study, there were 299 Bovine Leukocyte Antigen (BoLA)-DRB3 alleles loaded onto the NetBoLAIIpan website. Pairwise differentiation of the BoLA-DRB3 alleles removed 4 that were identical in amino acid sequence (Supplemental File 1).
The remaining 295 MHC Class II alleles were tested for interaction with the previously defined conserved C. burnetii proteome consisting of 1,022 proteins [29]. These proteins were broken into 15-mer peptides, resulting in 293,520 peptides that were tested. Of these peptides, 55.8% were predicted to interact with at least one MHC Class II allele. However, only 6,155 peptides were considered of interest because of strong binding predicted with 45% of the alleles (133) or because of either strong or weak binding predicted with 90% of the alleles (266). One-hundred fifty peptides were predicted to bind either strongly or weakly with all the alleles tested (Supplemental Table 5). Three peptides were predicted to bind strongly with 99% of the cattle alleles (Table 1); however, the start position of the 15-mer peptides of the protein PanC differ by only one amino acid, 240 and 241, and so, are likely comprised of the same T-cell epitope.
|
Position |
Peptide |
GenbankID |
SB |
WB |
TB |
Gene Name |
Locus Tag |
Location |
|
240 |
VGDIRLIDNIPFAKD |
AAO89975.1 |
292 |
3 |
295 |
panC |
CBU_0423 |
CYTOPLASM |
|
241 |
GDIRLIDNIPFAKDK |
AAO89975.1 |
292 |
3 |
295 |
panC |
CBU_0423 |
CYTOPLASM |
|
252 |
DSYLKYAPIHAVGAP |
AAO91502.1 |
292 |
3 |
295 |
|
CBU_2013 |
CYTOPLASM |
|
Position indicates the start of the peptide within the protein of interest, where the first amino acid in a protein is labeled position 1. Genbank ID, gene name, and locus tag represent protein information obtained from NCBI in the Nine Mile RSA 493 assembly. Protein location was previously predicted using the Inmembrane program [67]. The overlapping amino acids of the PanC peptides are indicated in bold and underlined. Abbreviations: SB: Strong Binders; WB: Weak Binders; TB: Total Binders. |
||||||||
Comparison of ruminant DRA proteins
The MHC Class II molecule is composed of two glycoproteins, the alpha and beta chains [4,26,35], which together form a peptide binding groove [1,26]. The alpha chains (DRA proteins) tend to be monomorphic whereas the beta chains (DRB proteins) are highly polymorphic [53-56,59,60,62,65]. Designed as a bioinformatic tool for cattle, the NetBoLAIIpan program holds the DRA protein constant but allows the user to choose the DRB protein against which peptides can be assessed for binding. However, the phylogenetic relationship between cattle and small ruminants is close and is reflected in the high sequence identity of cattle, sheep, and goat DRAs (Figure 1). As calculated using the BLASTp, the sheep and goat DRA proteins respectively share 96.8% and 96.1% identity with the cattle DRA protein. When comparing only the alpha 1 domain of DRA, that is the portion of DRA that participates in forming the peptide groove [26,56,66], the sheep and goat sequences respectively share 96.4% and 94.1% identity with cattle DRA (underlined and bolded in Figure 1). As a reference, the human DRA protein (GenbankID P01903.2) shares 80.71% identity with cattle DRA. Thus, despite the software limitation, we anticipate the pairing of small ruminant DRB proteins with the cattle DRA protein predicts T-cell epitopes that are robust to these few DRA differences between these species.
Figure 1. DRA protein alignment within ruminant species of interest. Alignment of the DRA protein from cattle, goats, and sheep [38,52,56]. The identifier in parenthesis is the Genbank ID for the aligned protein. Each cell contains an amino acid or a decimal which indicates no change from the cattle DRA reference protein. The bolded and underlined amino acids within the reference protein mark the alpha 1 domain of the protein.
T-cell epitopes predicted for MHC Class II alleles of sheep and goat
The sheep DRB alleles located on the Immuno Polymorphism Database (IPD)-MHC database have been identified within breeds prominent in the United States, including the Suffolk, Rambouillet, Polypay, Columbia, and Texel (Supplemental Table 2) [68]. In contrast, many of the MHC Class II DRB goat alleles are from the Scottish Cashmere goat, an infrequent breed within the United States [37]. Therefore, the whole genome sequences present on the National Center for Biotechnology Information (NCBI) database for Saanen and San Clemente goat breeds were examined for MHC Class II DRB alleles within chromosome 23. As shown in Supplemental Table 3 and Supplemental File 1, two alleles aligned with previously defined proteins and one de novo allele was established.
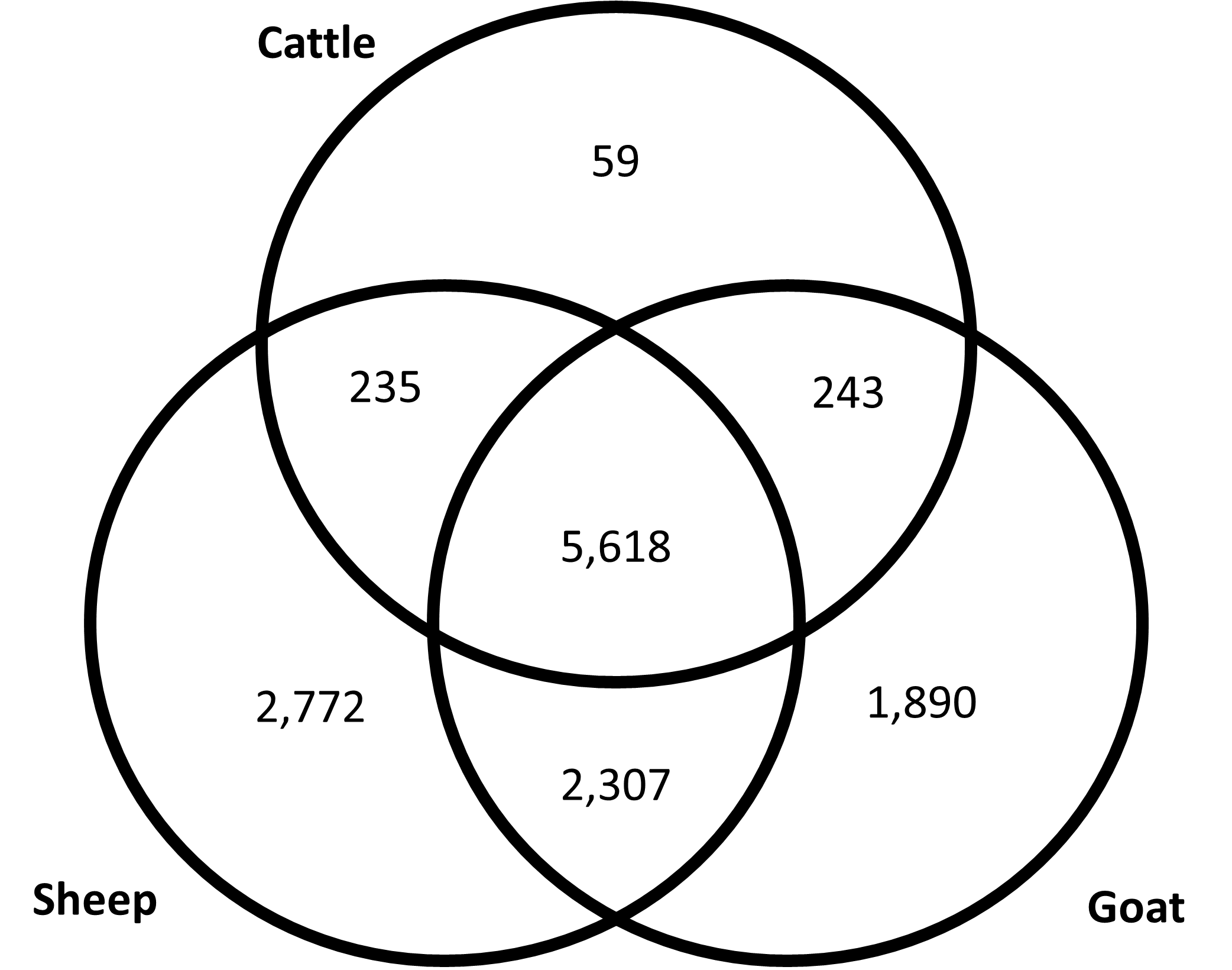
After training the software on 200,000 randomly generated peptides, the strength of interactions of each MHC Class II allele from sheep (21 DRB alleles) and goat (27 DRB alleles) with the conserved C. burnetii proteome [29] were predicted. There were 10,932 peptides predicted to interact with sheep alleles and 10,058 peptides predicted to interact with goat alleles. There were 5,618 high scoring peptides predicted to interact with MHC Class II molecules of sheep, goat, and cattle (Figure 2). This list reduced to 2,389 predicted MHC Class II T-cell epitopes for all three species when peptides overlapping by 7 amino acids were removed; this list comprises 0.8% of all peptides examined (Supplemental Table 6).
Figure 2. Predicted peptide interaction numbers for ruminant MHC Class II molecules. Venn Diagram depicting the number of predicted MHC Class II T-cell epitopes within each ruminant species and if peptides were predicted to interact with multiple species.
Pan-allelic peptides represent those predicted to bind all alleles tested. Pan-allelic strong binding was predicted for 287 peptides in sheep and 54 peptides in goats, 49 of which were pan-allelic strong binders in both sheep and goats. In including peptides predicted to strongly bind 95% of the cattle alleles (280 out of 295), eleven peptides were predicted to bind strongly with a great majority of the MHC Class II alleles tested across these three ruminant species (Table 2). Notably, each of these peptides are part of proteins likely to be localized to the bacterial cytoplasm.
|
Position |
Peptide |
GenbankID |
Gene Name |
Locus Tag |
Location |
|
103 |
AAKKIIKLIKDQKLK |
AAO89678.1 |
yajQ |
CBU_0114 |
CYTOPLASM |
|
105 |
IKRVKIFAAGKLEKP |
AAO89815.1 |
rplO |
CBU_0257 |
CYTOPLASM |
|
240 |
VGDIRLIDNIPFAKD |
AAO89975.1 |
panC |
CBU_0423 |
CYTOPLASM |
|
37 |
PVGFEIAQALHLPLD |
AAO90019.1 |
CBU_0470 |
CYTOPLASM |
|
|
31 |
DKSLVAIKNVTVNEP |
AAO90158.1 |
fabZ |
CBU_0614 |
CYTOPLASM |
|
275 |
ISSFVPIEIHVIPER |
AAO90229.1 |
CBU_0685 |
CYTOPLASM |
|
|
104 |
GEPFKYVVNFEVYPE |
AAO90277.1 |
tig |
CBU_0737 |
CYTOPLASM |
|
56 |
LSTIFIAVPIHHFKN |
AAO90505.1 |
CBU_0984 |
CYTOPLASM |
|
|
581 |
DIPFHAVEIEKLAHR |
AAO90739.1 |
CBU_1230 |
CYTOPLASM |
|
|
252 |
DSYLKYAPIHAVGAP |
AAO91502.1 |
CBU_2013 |
CYTOPLASM |
|
|
108 |
YDSIIKFAKANKLRI |
ACI15237.1 |
CBU_0095a |
CYTOPLASM |
|
|
* Strong binding predicted for 292 of 295 known DR cattle alleles and all tested alleles for sheep and goats. GenbankID, gene name, and locus tag annotations come from NCBI, where location was predicted by Inmembrane. The peptide and position are generated by NetBoLaIIpan. |
|||||
Comparison of MHC Class II T-cell epitopes between ruminant, human, and mouse species
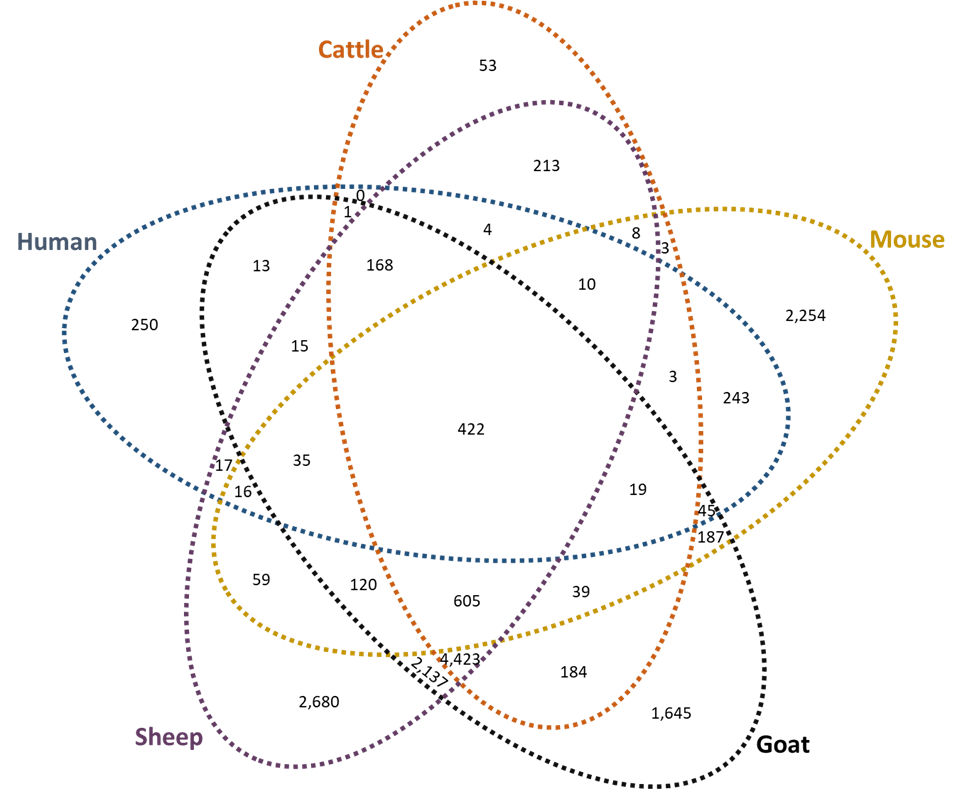
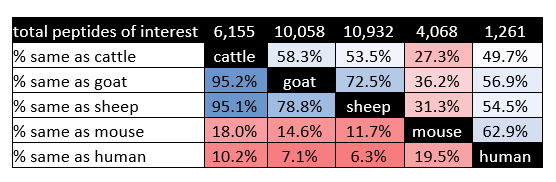
All data from the present and prior [29] studies are maintained on the same SQL Server Management Studio (SSMS) data server, which allowed us to readily compare T-cell epitope predictions across species. Peptides with exceptional binding characteristics for mouse and human MHC alleles were previously identified [29]. Similarly, for cattle, sheep, and goat MHC alleles, peptides were identified as having exceptional binding characteristics if predicted to strongly bind 45% or at least weakly bind 90% of the alleles tested within one of these species. This returned 15,871 peptides with exceptional within-species binding characteristics, the between species intersections of which are shown in Figure 3A. It is interesting to note that the largest number of peptides bound either only sheep, only goat, only mouse, sheep and goat, or sheep and goat and cattle alleles. It must be recognized that the criterion for making such distinctions is subject to the predominance of alleles in the populations of interest. This most likely affects the distinctions as applied to the relatively few alleles yet known for small ruminants and the eight alleles encompassed by NetMHCIIpan for mice. The increased peptide numbers predicted to bind all ruminant species supports their close evolutionary relationship. Figure 3B depicts the percent of peptides which remain consistent during direct species to species comparison, where ruminants share predicted interactions of greater than 50% of peptides in all cases. Nonetheless, there were 422 peptides identified with exceptional MHC allelic coverage for all five species, 256 of which differed by at least 50% in linear sequence identity (Supplemental Table 7).
3A
3B
Figure 3. Predicted peptide interaction numbers for MHC Class II molecules from all species. (A) Peptide numbers which reached high scoring cut-offs within each species (human, mouse, cattle, goat, and sheep) of interest for MHC Class II allelic interaction. The Venn Diagram breaks out the predicted peptides by how they interacted between species. (B) Percent of peptides predicted to interact between species in a direct species to species comparison. The top row indicates the total number of high scoring predicted MHC Class II peptide interactions per species. Percentage is color coded for ease, where blue is a high number of predicted peptides between species and red is the opposite.
Analysis of the pan-allelic T-cell epitopes predicted to interact with sheep, goat, and human species was decided upon due to human health being less affected by cattle exposure, following introduction of milk pasteurization [69], and since mice are less important to vaccine development for sheep, goats, and humans. Peptides which bound all goat or sheep alleles strongly or which are predicted to interact with all human alleles were filtered out of the data set (Table 3). As can be seen, 4 of the 11 T-cell epitopes described in Table 2 are present in the resultant dataset from the small ruminant species. In contrast to this, when assessing peptides which interacted with all human alleles tested, there were none that overlapped with high scoring ruminant peptides (Table 2 versus Table 3). However, two peptides remained consistent when assessing the dataset for human and small ruminant T-cell epitopes that interacted with all alleles tested. Filtering the data with these criteria returned proteins that localized beyond the cytoplasm within the membrane or that are secreted.
|
Position |
Peptide |
GenbankID |
Species |
SB |
WB |
TB |
Gene Name |
Locus Tag |
Location |
|
Peptides binding all small ruminant MHC Class II alleles |
|||||||||
|
105 |
KKIIKLIKDQKLKVQ |
AAO89678.1 |
Cow |
288 |
7 |
295 |
yajQ |
CBU_0114 |
CYTOPLASM |
|
Human |
102 |
62 |
164 |
||||||
|
Mouse |
5 |
0 |
5 |
||||||
|
152 |
KKNFYQLPINKVIDR |
AAO89725.1 |
Cow |
246 |
41 |
287 |
CBU_0165 |
Membrane |
|
|
Human |
98 |
47 |
145 |
||||||
|
Mouse |
4 |
4 |
8 |
||||||
|
197 |
QMEFTAIRINPVVAG |
AAO90399.1 |
Cow |
271 |
22 |
293 |
CBU_0866 |
MEMBRANE |
|
|
Human |
119 |
79 |
198 |
||||||
|
Mouse |
7 |
1 |
8 |
||||||
|
9 |
DKEIRAISDYVVNHK |
AAO90441.1 |
Cow |
271 |
21 |
292 |
prpD |
CBU_0912 |
CYTOPLASM |
|
Human |
171 |
35 |
206 |
||||||
|
Mouse |
6 |
1 |
7 |
||||||
|
235 |
RADMFIAVHADAYKN |
AAO90598.2 |
Cow |
250 |
42 |
292 |
CBU_1085 |
CYTOPLASM |
|
|
Human |
84 |
101 |
185 |
||||||
|
Mouse |
4 |
4 |
8 |
||||||
|
581 |
DIPFHAVEIEKLAHR |
AAO90739.1 |
Cow |
286 |
7 |
293 |
CBU_1230 |
CYTOPLASM |
|
|
Human |
157 |
36 |
193 |
||||||
|
Mouse |
7 |
1 |
8 |
||||||
|
255 |
DSDFIAFLKNQAADE |
AAO90852.2 |
Cow |
278 |
15 |
293 |
CBU_1349 |
CYTOPLASM |
|
|
Human |
121 |
60 |
181 |
||||||
|
Mouse |
5 |
2 |
7 |
||||||
|
306 |
DNGYRSIHTAVIGPE |
AAO90878.1 |
Cow |
278 |
15 |
293 |
relA |
CBU_1375 |
CYTOPLASM |
|
Human |
190 |
15 |
205 |
||||||
|
Mouse |
8 |
0 |
8 |
||||||
|
205 |
QGEYIIDIAEALKAK |
AAO91497.1 |
Cow |
267 |
28 |
295 |
argS |
CBU_2008 |
CYTOPLASM |
|
Human |
145 |
61 |
206 |
||||||
|
Mouse |
5 |
3 |
8 |
||||||
|
252 |
DSYLKYAPIHAVGAP |
AAO91502.1 |
Cow |
292 |
3 |
295 |
CBU_2013 |
CYTOPLASM |
|
|
Human |
137 |
66 |
203 |
||||||
|
Mouse |
8 |
0 |
8 |
||||||
|
108 |
YDSIIKFAKANKLRI |
ACI15237.1 |
Cow |
286 |
9 |
295 |
CBU_0095a |
CYTOPLASM |
|
|
Human |
153 |
52 |
205 |
||||||
|
Mouse |
7 |
1 |
8 |
||||||
|
Peptides binding all human MHC Class II alleles |
|||||||||
|
226 |
EGAIRHTHVIPIAGD |
AAO89704.2 |
Goat |
24 |
3 |
27 |
ftsA
|
CBU_0140
|
CYTOPLASM
|
|
Sheep |
20 |
1 |
21 |
||||||
|
Cow |
280 |
15 |
295 |
||||||
|
Mouse |
7 |
1 |
8 |
||||||
|
259 |
QIKIKYASVLPEEVN |
AAO89704.2 |
Goat |
25 |
2 |
27 |
ftsA
|
CBU_0140
|
CYTOPLASM
|
|
Sheep |
19 |
2 |
21 |
||||||
|
Cow |
273 |
22 |
295 |
||||||
|
Mouse |
7 |
1 |
8 |
||||||
|
9 |
DKEIRAISDYVVNHK |
AAO90441.1 |
Goat |
27 |
0 |
27 |
prpD |
CBU_0912 |
CYTOPLASM |
|
Sheep |
21 |
0 |
21 |
||||||
|
Cow |
271 |
21 |
292 |
||||||
|
Mouse |
6 |
1 |
7 |
||||||
|
169 |
RQSIRYYHTAAAIKN |
AAO90977.1 |
Goat |
19 |
8 |
27 |
|
CBU_1480 |
Membrane |
|
Sheep |
17 |
4 |
21 |
||||||
|
Cow |
213 |
80 |
293 |
||||||
|
Mouse |
7 |
1 |
8 |
||||||
|
390 |
RGKFKIYIADPAIAP |
AAO91005.1 |
Goat |
25 |
2 |
27 |
|
CBU_1508 |
CYTOPLASM |
|
Sheep |
21 |
0 |
21 |
||||||
|
Cow |
232 |
59 |
291 |
||||||
|
Mouse |
6 |
2 |
8 |
||||||
|
678 |
GNKIIQIAPARVANR |
AAO91357.1 |
Goat |
25 |
2 |
27 |
parC |
CBU_1866 |
CYTOPLASM |
|
Sheep |
17 |
4 |
21 |
||||||
|
Cow |
273 |
22 |
295 |
||||||
|
Mouse |
7 |
1 |
8 |
||||||
|
403 |
RQNIRAVDTQQVTAA |
AAO91392.1 |
Goat |
18 |
9 |
27 |
|
CBU_1901 |
Secreted |
|
Sheep |
14 |
7 |
21 |
||||||
|
Cow |
206 |
87 |
293 |
||||||
|
Mouse |
7 |
1 |
8 |
||||||
|
329 |
NNAIRYAKNVNVRIQ |
AAO91494.1 |
Goat |
25 |
2 |
27 |
rstB |
CBU_2005 |
Membrane |
|
Sheep |
21 |
0 |
21 |
||||||
|
Cow |
283 |
12 |
295 |
||||||
|
Mouse |
6 |
2 |
8 |
||||||
|
205 |
QGEYIIDIAEALKAK |
AAO91497.1 |
Goat |
27 |
0 |
27 |
argS |
CBU_2008 |
CYTOPLASM |
|
Sheep |
21 |
0 |
21 |
||||||
|
Cow |
267 |
28 |
295 |
||||||
|
Mouse |
5 |
3 |
8 |
||||||
|
T-cell epitopes predicted to bind all small ruminant alleles are above and epitopes predicted to bind all human alleles are below. The number of alleles each peptide interacted within other species either strongly (SB), weakly (WB), or in totality (TB) are notated. Bolded rows indicate that the predicted peptide interacted with all small ruminant and human alleles tested and underlined rows indicate peptides previously returned in Table 2. GenbankID, gene name, and locus tag are defined by NCBI. Inmembrane was used to predict protein location, where a potentially surface exposed protein is lowercase. Position and peptide are outputs from the NetBolaIIpan bioinformatic program. |
|||||||||
T-cell epitopes predicted for MHC Class I alleles of sheep and goats
Unlike the MHC Class II mature molecule using two proteins to produce the peptide binding groove, the MHC Class I peptide binding groove is only made from one protein [26]. For MHC Class I T-cell epitope prediction, the NetMHCpan program was used [28]. Similar to NetBoLAIIpan, the program was built initially to predict T-cell epitopes in the cattle species. Therefore, 160,000 randomly generated peptides were tested for each allele run on the program to generate a rank score to enable C. burnetii proteome analysis.
NetMHCpan breaks each protein into consecutive 8-, 9-, 10-, and 11-mers producing 1,196,551 peptides for testing from the conserved 1,002 C. burnetii proteins. Following loading of bioinformatic results into the SSMS database, it was calculated that 9,581 and 12,669 peptides for sheep and goat respectively reached the threshold of interacting with 60% of the species alleles tested or interacting strongly with 45% of the species alleles tested. Initial analysis of this data determined that 14 peptides were predicted to interact with all 56 sheep alleles strongly and that 25 peptides were predicted to interact with all 18 goat alleles strongly (Table 4).
|
Position |
Peptide |
GenbankID |
Gene Name |
Locus Tag |
Location |
|
Predicted strong binding pan-allelic peptides predicted for sheep MHC Class I presentation (56 alleles tested) |
|||||
|
302 |
VTYPPPKTL |
AAO89834.2 |
CBU_0276 |
CYTOPLASM |
|
|
333 |
RAYDVSFSL |
AAO89886.2 |
thiC |
CBU_0330 |
CYTOPLASM |
|
172 |
SSYPHPFLM |
AAO90011.1 |
pdhA |
CBU_0461 |
CYTOPLASM |
|
242 |
TVYPKTHYV |
AAO90064.1 |
uvrB |
CBU_0518 |
CYTOPLASM |
|
114 |
RAYEAIQSL |
AAO90172.1 |
ppa |
CBU_0628 |
CYTOPLASM |
|
67 |
AAYTSPHLL |
AAO90423.1 |
folC |
CBU_0894 |
CYTOPLASM |
|
196 |
SVYEQVHLL |
AAO90423.1 |
folC |
CBU_0894 |
CYTOPLASM |
|
302 |
YTFEKQLVL |
AAO90655.1 |
secD |
CBU_1142 |
Membrane |
|
482 |
SVYEGIPHL |
AAO90700.2 |
ftsK |
CBU_1191 |
Membrane |
|
335 |
FTYPKVPNL |
AAO90866.1 |
CBU_1363 |
CYTOPLASM |
|
|
126 |
SAFEWHLTF |
AAO90883.1 |
CBU_1380 |
MEMBRANE |
|
|
377 |
FSSPIFHEF |
AAO91209.1 |
CBU_1714 |
CYTOPLASM |
|
|
54 |
HTFPGVIQL |
AAO91456.1 |
CBU_1967 |
MEMBRANE |
|
|
64 |
IAAPLPIQL |
ACI15273.1 |
CBU_1067a |
Membrane |
|
|
Predicted strong binding pan-allelic peptides for goat MHC Class I presentation (18 alleles tested) |
|||||
|
273 |
IQYQGPILL |
AAO89579.1 |
dacB |
CBU_0009 |
Secreted |
|
6 |
MQVPLQITL |
AAO89590.2 |
|
CBU_0020 |
CYTOPLASM |
|
136 |
YQSEVQKEL |
AAO89696.1 |
ftsW |
CBU_0132 |
MEMBRANE |
|
33 |
AQSPLLHYL |
AAO89719.1 |
pilB |
CBU_0155 |
CYTOPLASM |
|
132 |
FQIKPPHQL |
AAO89740.2 |
|
CBU_0180 |
Cell Wall |
|
302 |
VTYPPPKTL |
AAO89834.2 |
|
CBU_0276 |
CYTOPLASM |
|
101 |
YQYDNVRSV |
AAO89868.2 |
|
CBU_0311 |
Membrane |
|
18 |
AQYPSPQLM |
AAO89889.1 |
thiG |
CBU_0333 |
CYTOPLASM |
|
114 |
YQIELIIEL |
AAO89932.1 |
ampD |
CBU_0379 |
Cell Wall |
|
119 |
KQADIYPTL |
AAO89970.1 |
|
CBU_0418 |
CYTOPLASM |
|
172 |
SSYPHPFLM |
AAO90011.1 |
pdhA |
CBU_0461 |
CYTOPLASM |
|
39 |
QQLEPSVTL |
AAO90063.2 |
aspB |
CBU_0517 |
CYTOPLASM |
|
220 |
YQKERVLTF |
AAO90095.2 |
rodA |
CBU_0549 |
MEMBRANE |
|
10 |
TQFEDLPSL |
AAO90111.1 |
|
CBU_0567 |
CYTOPLASM |
|
114 |
RAYEAIQSL |
AAO90172.1 |
ppa |
CBU_0628 |
CYTOPLASM |
|
77 |
FQFTRPHYL |
AAO90288.1 |
|
CBU_0748 |
Lipoprotein |
|
67 |
AAYTSPHLL |
AAO90423.1 |
folC |
CBU_0894 |
CYTOPLASM |
|
126 |
SQLPVIQKL |
AAO90606.1 |
|
CBU_1093 |
MEMBRANE |
|
166 |
HQNTPIQQL |
AAO90644.1 |
|
CBU_1131 |
CYTOPLASM |
|
482 |
SVYEGIPHL |
AAO90700.2 |
ftsK |
CBU_1191 |
Membrane |
|
62 |
FQLEHAHFL |
AAO90820.1 |
|
CBU_1316 |
CYTOPLASM |
|
513 |
SQQEKTIQL |
AAO91182.1 |
|
CBU_1686 |
CYTOPLASM |
|
156 |
SQNPALHAL |
AAO91229.2 |
|
CBU_1735 |
Membrane |
|
252 |
SSAPHTHAL |
AAO91476.1 |
apaH |
CBU_1987 |
CYTOPLASM |
|
38 |
YQFPQAPNM |
AAO91509.2 |
|
CBU_2021 |
CYTOPLASM |
|
Pan-allelic strong scoring peptide interactions for sheep are above and those for goats are below. Bolded and underlined rows indicate predicted T-cell epitopes that remained the same between these small ruminant species. Position and peptide are generated by NetMHCpan, where the Genbank ID, gene name, and locus tag are from NCBI. Location is estimated by the Inmembrane program; lowercase locations indicate the potential for surface exposure. |
|||||
Prediction of C. burnetii proteins which interact with cattle MHC Class I alleles has been presented previously [29]. Therefore, high scoring predicted T-cell epitopes were compared between all three ruminant species. Initially, this resulted in return of 3,547 peptides of interest, where this number dropped to 3,357 peptides when those overlapping by fifty percent, or more, were removed (Supplemental Table 8). Further filtering the data to isolate predicted T-cell epitopes that either bound all small ruminant alleles strongly or interacted with 98% of cattle alleles returned one peptide of interest. This peptide is present in Table 4 for locus tag CBU_0628 at position 114.
MHC Class I T-cell epitopes predicted in all five species
Similar to cattle MHC Class I allele T-cell epitope prediction, mouse and human prediction for MHC Class I alleles has been published previously [29]. Comparison of high scoring T-cell epitopes between all five species (goat, sheep, cattle, human, and mouse) returned 766 predicted T-cell epitopes (Supplemental Table 9). As done for MHC Class II predicted T-cell epitopes, primary attention was given to the small ruminant and human species. When assessing for peptides predicted to strongly bind with all the small ruminant alleles tested, there were no additions or subtraction from the bolded peptides in Table 4. Prior isolation of predicted T-cell epitopes that interacted with 90% of the tested human alleles, determined that there were 3 peptides of interest [29]. Of these, two peptides recurred in the present study (Table 5). The scores of the peptides as they relate to the other species tested are also present within Table 5, where the predicted T-cell epitopes are near the cut-offs given for exceptionally high interaction within the alternate species tested.
|
Position |
Peptide |
GenbankID |
Species |
SB |
WB |
TB |
Gene Name |
Locus Tag |
Location |
|
54 |
HTFPGVIQL |
AAO91456.1 |
Sheep |
56 |
0 |
56 |
CBU_1967 |
MEMBRANE |
|
|
Mouse |
3 |
4 |
7 |
||||||
|
Goat |
17 |
1 |
18 |
||||||
|
Cow |
75 |
26 |
101 |
||||||
|
113 |
ATYGHIHQM |
AAO91555.1 |
Sheep |
54 |
2 |
56 |
CBU_2071 |
MEMBRANE |
|
|
Mouse |
5 |
2 |
7 |
||||||
|
Goat |
17 |
1 |
18 |
||||||
|
Cow |
87 |
15 |
102 |
||||||
|
GenbankID, gene name, and locus tag are as notated on NCBI under the Nine Mile RSA493 assembly. Position and peptide are designated by NetMHCpan, while location was previously predicted by Inmembrane. Species tested beyond humans have their scores for each peptide of interest. The calculated scores are for strong binders (SB), weak binders (WB), and total binders (TB). |
|||||||||
T-cell epitope dense proteins
Epitope dense proteins have been previously defined as proteins which contain multiple peptides of interest [29,70,71]. Prior evaluation of epitope dense proteins from C. burnetii was completed when assessing human, mouse, and cattle species [29]. Re-examination of the data following the addition of goat and sheep species resulted in a list of predominantly the same proteins of interest (Supplemental Table 10). Importantly there were 14 MHC Class II proteins and 3 MHC Class I proteins which no longer achieved the set cut-off values. Furthermore, there was one novel protein for MHC Class II, AAO90920.1 or radA, returned.
Comparison of MHC Class I and Class II predicted T-cell epitopes
In analyzing both MHC Class I and Class II allele types for predicted T-cell epitopes, we are able to evaluate for epitope dense proteins and for overlapping epitopes that fulfill both classes of MHC allele. Resultant data from all 5 species with overlapping peptides removed were combined for MHC Class II and MHC Class I. Afterwards, any protein suggested to contain at least two T-cell epitopes were isolated for further examination (Supplemental Table 11).
Recent experimentation indicated that surface isolated proteins from Nine Mile Phase II bacteria produced a protective immune response in mice and guinea pigs [72]. Furthermore, these isolated proteins lowered the clinical disease seen in non-human primates, where vaccinated animals did not become febrile or have an increased respiratory rate [72]. This data place emphasis on surface localized proteins; therefore, membrane localized or secreted proteins were isolated and are presented in Table 6.
|
Genbank ID |
MHC Class I |
MHC Class II |
Epitope Dense |
Gene Name |
Locus Tag |
Location |
|
AAO89616.1* |
312 |
257, 338 |
No |
CBU_0049 |
Membrane |
|
|
AAO89682.2* |
348 |
426, 508 |
No |
ftsI |
CBU_0118 |
Membrane |
|
AAO89683.2 |
175 |
342 |
No |
CBU_0119 |
Membrane |
|
|
AAO89702.1* |
19 |
214 |
No |
ftsQ |
CBU_0138 |
Membrane |
|
AAO89725.1* |
87 |
152 |
No |
CBU_0165 |
Membrane |
|
|
AAO89757.1 |
96, 147, 165, 375, 637, 755 |
748 |
Yes |
CBU_0197 |
Membrane |
|
|
AAO89774.2 |
38, 60, 355, 421, 488, 509 |
283 |
Yes |
CBU_0215 |
Cellwall |
|
|
AAO89870.2* |
59 |
400 |
No |
CBU_0313 |
Cellwall |
|
|
AAO89926.1* |
146, 291 |
302 |
No |
CBU_0372 |
Membrane |
|
|
AAO89978.2* |
26, 211 |
59 |
No |
CBU_0426 |
MEMBRANE |
|
|
AAO90003.2* |
3 |
49 |
No |
fimT |
CBU_0453 |
Membrane |
|
AAO90031.1 |
49 |
246 |
No |
CBU_0482 |
Secreted |
|
|
AAO90110.1* |
199 |
320 |
No |
CBU_0566 |
MEMBRANE |
|
|
AAO90155.1 |
470, 551, 795 |
257, 320 |
Yes |
yaeT |
CBU_0611 |
Membrane |
|
AAO90276.1* |
173 |
89 |
No |
CBU_0736 |
Secreted |
|
|
AAO90399.1* |
63, 152 |
197 |
No |
CBU_0866 |
MEMBRANE |
|
|
AAO90424.2* |
47, 132 |
190 |
No |
dedD |
CBU_0895 |
Membrane |
|
AAO90487.1* |
182, 304, 325 |
408 |
No |
cydA-2 |
CBU_0965 |
Membrane |
|
AAO90508.2* |
184 |
62 |
No |
CBU_0987 |
Lipoprotein |
|
|
AAO90522.1* |
265 |
396 |
No |
lolC |
CBU_1001 |
MEMBRANE |
|
AAO90656.1* |
81 |
8 |
No |
yajC |
CBU_1143 |
MEMBRANE |
|
AAO90684.1 |
26, 222 |
208 |
No |
CBU_1175 |
MEMBRANE |
|
|
AAO90696.1 |
59 |
48, 197 |
No |
CBU_1187 |
Secreted |
|
|
AAO90700.2* |
16, 288, 482 |
244 |
No |
ftsK |
CBU_1191 |
Membrane |
|
AAO90703.1* |
158, 241 |
166 |
No |
CBU_1194 |
Membrane |
|
|
AAO90737.1 |
150, 257, 419, 468 |
254 |
Yes |
qseC |
CBU_1228 |
Membrane |
|
AAO90753.1* |
117, 378, 431 |
427 |
No |
CBU_1244 |
MEMBRANE |
|
|
AAO90948.1* |
294 |
73 |
No |
CBU_1451 |
Membrane |
|
|
AAO90965.2 |
109, 236, 367, 952 |
168, 608, 699 |
Yes |
CBU_1468 |
Membrane |
|
|
AAO90977.1* |
8 |
169 |
No |
CBU_1480 |
Membrane |
|
|
AAO91047.2 |
236, 462 |
721 |
No |
ptsP |
CBU_1550 |
Membrane |
|
AAO91053.1* |
107 |
90 |
No |
CBU_1556 |
MEMBRANE |
|
|
AAO91093.1* |
334, 538 |
666 |
No |
rpoD |
CBU_1596 |
Secreted |
|
AAO91122.1* |
179 |
18 |
No |
icmG |
CBU_1626 |
Membrane |
|
AAO91140.1* |
10, 93 |
192 |
No |
dotC |
CBU_1644 |
Lipoprotein |
|
AAO91155.2 |
66, 97, 182 |
59 |
No |
CBU_1659 |
MEMBRANE |
|
|
AAO91229.2 |
44, 156 |
31 |
No |
CBU_1735 |
Membrane |
|
|
AAO91235.1* |
39, 81 |
255 |
No |
CBU_1741 |
Lipoprotein |
|
|
AAO91260.1* |
509 |
254 |
No |
feoB |
CBU_1766 |
Membrane |
|
AAO91303.1 |
41 |
34, 85 |
No |
macA |
CBU_1810 |
Lipoprotein |
|
AAO91311.1* |
293 |
317 |
No |
CBU_1818 |
MEMBRANE |
|
|
AAO91329.1* |
57, 285 |
40 |
No |
CBU_1836 |
Membrane |
|
|
AAO91360.1* |
17 |
166 |
No |
CBU_1869 |
Secreted |
|
|
AAO91378.1 |
540, 553 |
103, 253 |
No |
ponA |
CBU_1887 |
Membrane |
|
AAO91393.1 |
105, 117, 287 |
365, 409 |
Yes |
CBU_1902 |
Secreted |
|
|
AAO91411.1* |
400, 532 |
185, 282 |
No |
yidC |
CBU_1920 |
MEMBRANE |
|
AAO91419.2 |
11, 298, 505, 668 |
594 |
Yes |
CBU_1928 |
Membrane |
|
|
AAO91467.1 |
164, 171, 616, 833 |
730 |
Yes |
ostA |
CBU_1978 |
Secreted |
|
AAO91474.1 |
125 |
121 |
No |
CBU_1985 |
Membrane |
|
|
AAO91516.1* |
92 |
16 |
No |
CBU_2029 |
Secreted |
|
|
AAO91543.1* |
320 |
139 |
No |
CBU_2058 |
MEMBRANE |
|
|
GenbankID, gene name, and locus tag are protein information isolated from NCBI from Nine Mile RSA493. MHC Class I and MHC Class II columns indicate the positions which are considered high scoring epitopes in all 5 species tested. A protein is considered epitope dense if it has greater than or equal to 5 predicted T-cell epitopes. Locations were previously predicted by the Inmembrane program, where a potentially surface exposed protein is notated by upper and lower case listed positions. Asterisks following the GenbankID denote newly predicted proteins of interest between this manuscript and the prior analysis completed with human, mice, and cattle only [29]. Overlapping MHC Class I and Class II epitopes have their positions underlined in their respective columns. |
||||||
Filtering the data in this way returned 51 proteins of interest. Notably, 33 of these proteins are newly identified as compared to the previous study predicting T-cell epitopes in humans, mouse, and cattle species, but lacked small ruminant examination [29]. This emphasizes the importance of including all species that are rational choices for developing a subunit vaccine. Within the present proteins, there are 10 which contain T-cell epitopes predicted to overlap between MHC Class I and MHC Class II surface molecules. These include CBU_0197, CBU_1175, CBU_1187, CBU_1194, CBU_1228 (qseC), CBU_1244, CBU_1659, CBU_1735, CBU_1810 (macA), and CBU_1985. Furthermore, it is intriguing to observe that only 19 of the proteins within this list contain gene names. And while some of these proteins may have been studied by direct means, it is more than likely that homology between proteins of alternate bacterial species has allowed for the identification of the presently designated genes.
Discussion
T-cell epitope prediction has expanded in the last few decades to include species beyond humans and common animal models, mice and swine for example [27,28]. This is beneficial when studying animal health related pathogens or pathogens that are responsible for zoonotic transmission. C. burnetii is one such pathogen that can result in Q fever during human exposure and may lead to reproductive related issues in ruminant species [11,12]. The present study employed current bioinformatic T-cell prediction programs to evaluate T-cell epitopes for three ruminant species. This required isolation of sheep and goat alleles, execution of bioinformatic programs, and assembling resultant data in a comprehensive fashion. A summary of the additional documents is available in Supplemental Table 12 to help the reader with file orientation.
After assessing the sheep and goat MHC alleles available for analysis, it was determined that the sheep species had a greater capacity to represent common North American breeds of interest. Notably, the IPD-MHC database maintains several sheep MHC alleles belonging to either Class I or Class II. Importantly, while many of the MHC Class II alleles have been directly sequenced from breeds of interest, the MHC Class I alleles have not been studied in depth. For this reason, whole genome sequences for sheep breeds of interest were isolated from NCBI and screened for additional MHC Class I sheep candidates. This increased the sheep MHC Class I alleles by twenty-four, of which both IPD-MHC and whole genome defined alleles were included in the present analysis. In contrast, assessing goat alleles present on the IPD-MHC database determined that further study is required to create an in-depth library of carried genes beyond the Scottish Cashmere breed [37]. Specifically, Ballingall et al. have presently sequenced twenty-eight MHC Class II alleles, of which two were identified when screening the Saanen and San Clemente genomes available on NCBI. Furthermore, there are no MHC Class I alleles at present for goat species on the IPD-MHC database and manuscript investigation only yielded one result [39]. Screening of the Saanen and San Clemente genomes produced seventeen MHC Class I alleles for testing in the present assay, but goat MHCs clearly require further definition.
Following allele definition, T-cell epitope predictions were run for the ruminant species and compared within the Bovidae family or between all five species of interest. The importance of interspecies comparison is demonstrated within recent C. burnetii research defining T-cell epitopes through T-cell recall responses against pathogen peptides [73-75]. Specifically, it was shown that several peptides could elicit IFN-γ secretion by human T-cells following natural exposure to C. burnetii during the 2007-2010 Netherlands outbreak [73,74]. A subset of the peptides which elicited a recall response were further tested within the mouse species, where it was determined that mouse T-cells had a recall response to only 33% of the tested peptides following C. burnetii inoculation [75]. Further testing within these experiments showed that mice predominantly responded to one of the twenty-seven tested peptides and that this response did not elicit protection during inoculation assays. A similar phenomenon can be seen herein, where 83 out of the 122 resultant proteins of interest returned in the present study were novel compared to the prior study (Supplemental Table 11) [29]. While these numbers represent the initial assessment of high scoring proteins of interest, the pattern continued when considering MHC Class-overlapping T-cell epitopes and epitope dense proteins of interest. Specifically, when assessing peptides of interest that overlapped between the MHC Class I and Class II allele presentation, there were two new proteins of interest, CBU_1194 and CBU_1244. Accompanying these two additional proteins, there were fifteen proteins no longer present from the original dataset (Supplemental Table 13) [29].
Due to the interspecies differences seen, testing of the present T-cell epitope predictions needs to be indicated by vaccine direction. Efforts to employ common C. burnetii proteins in vaccine development studies have occurred; however, subunit vaccine research has typically been focused on human health employing mouse models [73-76]. While safety and cost concerns continue to prohibit mass production of a formalin-inactivated whole cell vaccine within the United States, studies assessing these bacterins in other countries have permitted researchers a glimpse of protective phenotypes [15,17,77,78]. Future studies could choose to investigate proteins that are considered epitope dense or peptides which are suggested suitable to both MHC Class I and Class II presentation. While the current manuscript has not eliminated predicted epitopes of interest based on these distinctions, it has primarily focused on surface localized or secreted proteins. This is due to work by Gregory et al., which described surface isolated Nine Mile phase II proteins producing a protective immune response in both guinea pigs and mice [72].
It is important to mention that prior work promoting generation of a C. burnetii vaccine has suggested that multiple epitopes are required to produce a protective immune response [75,79-82]. For this reason, scientists have generated preliminary vaccines containing up to 28 epitopes of interest [75]. Keeping with this rationale would promote testing of overlapping MHC Class I and Class II T-cell epitopes, such as the 10 epitopes found in Table 6. Notably, only two of these proteins have a given name, MacA and QseC. Studies on either protein demonstrate the correct cellular location by Inmembrane, as they both contain transmembrane and periplasmic domains [35,83-86]. Direct study of the C. burnetii derived proteins has not yet occurred, but there is research from other Gram-negatives, like Escherichia coli. Specifically, MacA serves in a multimeric complex shown to export macrolides from inside the bacterium to the external environment [83,86]. In contrast, QseC is part of the quorum sensing cascade that can result in alternate transcription of virulence factors or motility components [84,85]. As can be seen, either protein’s activity is critical for bacterial virulence. Of the remaining eight overlapping MHC Class I and Class II epitopes, there are two new within this study due to the inclusion of small ruminants; comprised of CBU_1194 and CBU_1244. Even though these ten proteins represent a small subset of the overall proteome, there are a number of annotated hypothetical proteins which would benefit from further characterization. For example, recent examination of CBU_0307, which was previously described as containing a high scoring MHC Class I human T-cell epitope, has discerned the protein’s importance in membrane structural integrity [29,73,87].
Conclusion
Overall, this manuscript expands the scope of comparative predicted C. burnetii T-cell epitopes to include the small ruminant species. This represents one of the few manuscripts which has accounted for the cross-species C. burnetii pathogenesis during T-cell epitope prediction. During this analysis, it was determined that MHC allele characterization is required for the goat species. Nevertheless, novel proteins of interest were identified when considering the additional goat and sheep species. While T-cell epitopes of interest have been predicted, their ability to stimulate T-cells will be further elucidated by future work.
Declarations
Availability of data and materials
The previous dataset from the manuscript titled “Proteome-wide analysis of Coxiella burnetii for conserved T-cell epitopes with presentation across multiple host species,” is available on an Open Science Framework repository with accession number RN6QA (https://osf.io/rn6qa/). The present study has analyzed data within the published article, supplementary information, and a separate online repository. The datasets generated by this study are available in the National Agricultural Library Ag Data Commons through the Figshare portal under the title “Data for: Expansion of Proteome-wide C. burnetii Comparative T-Cell Epitope Prediction to Include Small Ruminant Hosts” (https://doi.org/10.15482/USDA.ADC.24712827.v1).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Funding
This work was supported by USDA-ARS 2090-32000-046-000D.
Acknowledgements
Not applicable.
Authors’ contributions
Conceptualization: LMWP, SNW; Methodology: LMWP, PCG, RK, DAS; Formal Analysis: LMWP; Writing-Original Draft: LMWP, PCG; Final Writing & Editing: LMWP, PCG, DAS, RK, SNW; Data curation: LMWP, RK, DAS. All authors read and approved the final manuscript.
References
2. Pollard AJ, Bijker EM. Publisher Correction: A guide to vaccinology: From basic principles to new developments. Nature Reviews Immunology. 2021 Feb;21(2):129.
3. Pollard AJ, Bijker EM. A guide to vaccinology: from basic principles to new developments. Nature Reviews Immunology. 2021 Feb;21(2):83-100.
4. Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, Rosales-Mendoza S. An overview of bioinformatics tools for epitope prediction: implications on vaccine development. Journal of Biomedical Informatics. 2015 Feb 1;53:405-14.
5. Krammer F. The human antibody response to influenza A virus infection and vaccination. Nature Reviews Immunology. 2019 Jun;19(6):383-97.
6. Seshadri R, Paulsen IT, Eisen JA, Read TD, Nelson KE, Nelson WC, et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proceedings of the National Academy of Sciences. 2003 Apr 29;100(9):5455-60.
7. Read AJ, Erickson S, Harmsen AG. Role of CD4+ and CD8+ T cells in clearance of primary pulmonary infection with Coxiella burnetii. Infection and Immunity. 2010 Jul;78(7):3019-26.
8. Hemsley CM, O’Neill PA, Essex-Lopresti A, Norville IH, Atkins TP, Titball RW. Extensive genome analysis of Coxiella burnetii reveals limited evolution within genomic groups. BMC Genomics. 2019 Dec;20(1):441.
9. Movahedi AR, Hampson DJ. New ways to identify novel bacterial antigens for vaccine development. Veterinary Microbiology. 2008 Sep 18;131(1-2):1-13.
10. Andoh M, Zhang G, Russell-Lodrigue KE, Shive HR, Weeks BR, Samuel JE. T cells are essential for bacterial clearance, and gamma interferon, tumor necrosis factor alpha, and B cells are crucial for disease development in Coxiella burnetii infection in mice. Infection and Immunity. 2007 Jul;75(7):3245-55.
11. Eldin C, Mélenotte C, Mediannikov O, Ghigo E, Million M, Edouard S, et al. From Q fever to Coxiella burnetii infection: a paradigm change. Clinical Microbiology Reviews. 2017 Jan;30(1):115-90.
12. Agerholm JS. Coxiella burnetii associated reproductive disorders in domestic animals-a critical review. Acta Veterinaria Scandinavica. 2013 Dec;55(1):13.
13. Maurin M, Raoult DF. Q fever. Clinical Microbiology Reviews. 1999 Oct 1;12(4):518-53.
14. Raoult D, Marrie TJ, Mege JL. Natural history and pathophysiology of Q fever. The Lancet Infectious Diseases. 2005 Apr 1;5(4):219-26.
15. Williams-Macdonald SE, Mitchell M, Frew D, Palarea-Albaladejo J, Ewing D, Golde WT, et al. Efficacy of Phase I and Phase II Coxiella burnetii Bacterin Vaccines in a Pregnant Ewe Challenge Model. Vaccines. 2023 Feb 22;11(3):511.
16. Van Moll P, Baumgärtner W, Eskens U, Hänichen T. Immunocytochemical demonstration of Coxiella burnetii antigen in the fetal placenta of naturally infected sheep and cattle. Journal of Comparative Pathology. 1993 Oct 1;109(3):295-301.
17. Arricau-Bouvery N, Souriau A, Bodier C, Dufour P, Rousset E, Rodolakis A. Effect of vaccination with phase I and phase II Coxiella burnetii vaccines in pregnant goats. Vaccine. 2005 Aug 15;23(35):4392-402.
18. Welsh HH, Lennette EH, Abinanti FR, Winn JF. Air-borne transmission of Q fever: the role of parturition in the generation of infective aerosols. Annals of the New York Academy of Sciences. 1958 Jun;70(3):528-40.
19. Heinzen RA, Hackstadt T, Samuel JE. Developmental biology of Coxiella burnetii. Trends in Microbiology. 1999 Apr 1;7(4):149-54.
20. McCAUL TF, Williams JC. Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. Journal of Bacteriology. 1981 Sep;147(3):1063-76.
21. Tigertt WD, Benenson AS, Gochenour WS. Airborne Q fever. Bacteriological Reviews. 1961 Sep;25(3):285-93.
22. Guatteo R, Seegers H, Taurel AF, Joly A, Beaudeau F. Prevalence of Coxiella burnetii infection in domestic ruminants: a critical review. Veterinary Microbiology. 2011 Apr 21;149(1-2):1-16.
23. McQuiston JH, Childs JE. Q fever in humans and animals in the United States. Vector Borne and Zoonotic Diseases. 2002 Sep 1;2(3):179-91.
24. O'Neill TJ. Coxiella burnetii in small ruminants and humans: Evidence-based control and prevention strategies with a model to assess financial impact of coxiellosis to the Ontario goat industry, in Population Medicine. 2012, University of Guelph: Guelph, Ontario.
25. Van Asseldonk MA, Prins J, Bergevoet RH. Economic assessment of Q fever in the Netherlands. Preventive Veterinary Medicine. 2013 Oct 1;112(1-2):27-34.
26. Sanchez-Trincado JL, Gomez-Perosanz M, Reche PA. Fundamentals and methods for T-and B-cell epitope prediction. Journal of Immunology Research. 2017 Oct;2017:2680160.
27. Fisch A, Reynisson B, Benedictus L, Nicastri A, Vasoya D, Morrison I, et al. Integral use of immunopeptidomics and immunoinformatics for the characterization of antigen presentation and rational identification of BoLA-DR–presented peptides and epitopes. The Journal of Immunology. 2021 May 15;206(10):2489-97.
28. Nielsen M, Connelley T, Ternette N. Improved prediction of bovine leucocyte antigens (BoLA) presented ligands by use of mass-spectrometry-determined ligand and in vitro binding data. Journal of Proteome Research. 2018 Jan 5;17(1):559-67.
29. Piel LM, Durfee CJ, White SN. Proteome-wide analysis of Coxiella burnetii for conserved T-cell epitopes with presentation across multiple host species. BMC bioinformatics. 2021 Jun 2;22(1):296.
30. Expasy RandSeq - Random protein sequence generator. [cited 2021; Available from: https://web.expasy.org/randseq/.
31. Mimotopes The Peptide Company Scrambled. [cited 2021; Available from: http://www.mimotopes.com/peptideLibraryScreening.asp?id=97.
32. Jørgensen KW, Buus S, Nielsen M. Structural properties of MHC class II ligands, implications for the prediction of MHC class II epitopes. PloS One. 2010 Dec 30;5(12):e15877.
33. Lundegaard C, Lund O, Buus S, Nielsen M. Major histocompatibility complex class I binding predictions as a tool in epitope discovery. Immunology. 2010 Jul;130(3):309-18.
34. Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Research. 2020 Jul 2;48(W1):W449-54.
35. Amills M, Ramiya V, Norimine J, Lewin HA. The major histocompatibility complex of ruminants. Revue Scientifique et Technique (International Office of Epizootics). 1998 Apr 1;17(1):108-20.
36. Miltiadou D, Ballingall KT, Ellis SA, Russell GC, McKeever DJ. Haplotype characterization of transcribed ovine major histocompatibility complex (MHC) class I genes. Immunogenetics. 2005 Aug;57:499-509.
37. Ballingall KT, Todd H. An official nomenclature for the major histocompatibility complex allele sequences from the domestic goat (Capra hircus). HLA. 2019 Jan;93(1):36-8.
38. Takada T, Kikkawa Y, Yonekawa H, Amano T. Analysis of goat MHC class II DRA and DRB genes: identification of the expressed gene and new DRB alleles. Immunogenetics. 1998 Oct;48:408-12.
39. Zidi A, Sànchez A, Obexer-Ruff G, Amills M. Sequence analysis of goat major histocompatibility complex class I genes. Journal of Dairy Science. 2008 Feb 1;91(2):814-7.
40. Siva Subramaniam N, Morgan EF, Wetherall JD, Stear MJ, Groth DM. A comprehensive mapping of the structure and gene organisation in the sheep MHC class I region. BMC Genomics. 2015 Dec;16:810.
41. Ballingall KT, Dicks K, Kyriazopoulou P, Herrmann-Hoesing L. Allelic nomenclature for the duplicated MHC class II DQ genes in sheep. Immunogenetics. 2019 Apr 1;71:347-51.
42. Ballingall KT, Tassi R. Sequence-based genotyping of the sheep MHC class II DRB1 locus. Immunogenetics. 2010 Jan;62:31-9.
43. Herrmann-Hoesing LM, White SN, Kappmeyer LS, Herndon DR, Knowles DP. Genomic analysis of Ovis aries (Ovar) MHC class IIa loci. Immunogenetics. 2008 Apr;60:167-76.
44. Herrmann-Hoesing LM, White SN, Mousel MR, Lewis GS, Knowles DP. Ovine progressive pneumonia provirus levels associate with breed and Ovar-DRB1. Immunogenetics. 2008 Dec;60:749-58.
45. Maccari G, Robinson J, Ballingall K, Guethlein LA, Grimholt U, Kaufman J, et al. IPD-MHC 2.0: an improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Research. 2017 Jan 4;45(D1):D860-4.
46. Atlija M, Gutíerrez-Gil B, Arranz JJ, Semmer J, Stear MJ, Buitkamp J. Major histocompatibility complex class IIB polymorphism in an ancient Spanish breed. Immunogenetics. 2015 Sep;67:531-7.
47. Dicks KL, Pemberton JM, Ballingall KT. Characterisation of major histocompatibility complex class IIa haplotypes in an island sheep population. Immunogenetics. 2019 May 1;71:383-93.
48. Fabb SA, Maddox JF, Gogolin-Ewens KJ, Baker L, Wu MJ, RBrandon M. Isolation, characterization and evolution of ovine major histocompatibility complex class II DRA and DQA genes. Animal Genetics. 1993 Aug;24(4):249-55.
49. Herrmann LM, Brown WC, Lewis GS, Knowles DP. Identification and phylogenetic analysis of 15 MHC class II DRB1 β1 expressed alleles in a ewe–lamb flock. Immunogenetics. 2005 Dec;57:855-63.
50. Konnai S, Nagaoka Y, Takeshima S, Onuma M, Aida Y. Sequences and diversity of 17 new Ovar‐DRB1 alleles from three breeds of sheep. European Journal of Immunogenetics. 2003 Aug;30(4):275-82.
51. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 2018 Jun;35(6):1547-9.
52. Ballingall KT, Rocchi MS, McKeever DJ, Wright F. Trans-species polymorphism and selection in the MHC class II DRA genes of domestic sheep. PLoS One. 2010 Jun 30;5(6):e11402.
53. Miyasaka T, Takeshima SN, Sentsui H, Aida Y. Identification and diversity of bovine major histocompatibility complex class II haplotypes in Japanese Black and Holstein cattle in Japan. Journal of Dairy Science. 2012 Jan 1;95(1):420-31.
54. Zhou H, Hickford JG, Fang Q, Byun SO. Identification of allelic variation at the bovine DRA locus by polymerase chain reaction-single strand conformational polymorphism. Journal of Dairy Science. 2007 Apr 1;90(4):1943-6.
55. Davies CJ, Joosten I, Andersson L, Arriens MA, Bernoco D, Bissumbhar B, et al. Polymorphism of bovine MHC class II genes. Joint report of the fifth international bovine lymphocyte antigen (BoLA) workshop, Interlaken, Switzerland, 1 august 1992. International Journal of Immunogenetics. 1994 Aug;21(4):259-89.
56. van der Poel JJ, Groenen MA, Dijkhof RJ, Ruyter D, Giphart MJ. The nucleotide sequence of the bovine MHC class II alpha genes: DRA, DQA, and DYA. Immunogenetics. 1990 Jan;31:29-36.
57. Ballingall KT, Ellis SA, MacHugh ND, Archibald SD, McKeever DJ. The DY genes of the cattle MHC: expression and comparative analysis of an unusual class II MHC gene pair. Immunogenetics. 2004 Feb;55:748-55.
58. Kumari N, Mishra SK, Saini S, Kumar A, Loat S, Dhilor N, et al. Identification of novel allelic patterns and evolutionary lineage of BoLA MHC class II DQA locus in indicine and taurine cattle. Animal Biotechnology. 2022 Dec 12;33(7):1746-52.
59. Maillard JC, Chantal I, Berthier D. Sequencing of four new BoLA-DRB3 and six new BoLA-DQB alleles. Animal Genetics. 2001 Feb;32(1):44-6.
60. Mikko S, Anderson L. Extensive MHC class II DRB3 diversity in African and European cattle. Immunogenetics. 1995 Sep;42:408-13.
61. Russell GC, Smith JA, Oliver RA. Structure of the BoLA-DRB3 gene and promoter. European Journal of Immunogenetics. 2004 Jun;31(3):145-51.
62. Sitte K, East IJ, Lavin MF, Jazwinska EC. Identification and characterization of new BoLA-DRB3 alleles by heteroduplex analysis and direct sequencing. Animal Genetics. 1995 Dec;26(6):413-7.
63. Nasir L, Ndiaye M, Seely C, Stear MJ. Sequence polymorphism in the bovine major histocompatibility complex DQB loci. Animal Genetics. 1997 Dec;28(6):441-5.
64. Norimine J, Brown WC. Intrahaplotype and interhaplotype pairing of bovine leukocyte antigen DQA and DQB molecules generate functional DQ molecules important for priming CD4+ T-lymphocyte responses. Immunogenetics. 2005 Nov;57:750-62.
65. Groenen MA, van der Poel JJ, Dijkhof RJ, Giphart MJ. The nucleotide sequence of bovine MHC class II DQB and DRB genes. Immunogenetics. 1990 Jan;31:37-44.
66. Behl JD, Verma NK, Tyagi N, Mishra P, Behl R, Joshi BK. The major histocompatibility complex in bovines: a review. International Scholarly Research Notices. 2012;2012:872710.
67. Perry AJ, Ho BK. Inmembrane, a bioinformatic workflow for annotation of bacterial cell-surface proteomes. Source Code for Biology and Medicine. 2013 Dec;8(1):9.
68. Thorne JW, Murdoch BM, Freking BA, Redden RR, Murphy TW, Taylor JB, et al. Evolution of the sheep industry and genetic research in the United States: opportunities for convergence in the twenty-first century. Animal Genetics. 2021 Aug;52(4):395-408.
69. Cerf O, Condron R. Coxiella burnetii and milk pasteurization: an early application of the precautionary principle?. Epidemiology & Infection. 2006 Oct;134(5):946-51.
70. Hisham Y, Ashhab Y. Identification of cross-protective potential antigens against pathogenic Brucella spp. through combining pan-genome analysis with reverse vaccinology. Journal of Immunology Research. 2018 Oct;2018:1474517.
71. Maman Y, Nir-Paz R, Louzoun Y. Bacteria modulate the CD8+ T cell epitope repertoire of host cytosol-exposed proteins to manipulate the host immune response. PLOS Computational Biology. 2011 Oct 13;7(10):e1002220.
72. Gregory AE, van Schaik EJ, Fratzke AP, Russell-Lodrigue KE, Farris CM, Samuel JE. Soluble antigens derived from Coxiella burnetii elicit protective immunity in three animal models without inducing hypersensitivity. Cell Reports Medicine. 2021 Dec 21;2(12):100461.
73. Scholzen A, Richard G, Moise L, Baeten LA, Reeves PM, Martin WD, et al. Promiscuous Coxiella burnetii CD4 epitope clusters associated with human recall responses are candidates for a novel T-cell targeted multi-epitope Q fever vaccine. Frontiers in immunology. 2019 Feb 15;10:207.
74. Scholzen A, Richard G, Moise L, Hartman E, Bleeker-Rovers CP, Reeves PM, et al. Coxiella burnetii epitope-specific T-cell responses in patients with chronic Q fever. Infection and Immunity. 2019 Oct;87(10):10-128.
75. Sluder AE, Raju Paul S, Moise L, Dold C, Richard G, Silva-Reyes L, Baeten LA, et al. Evaluation of a Human T Cell-Targeted Multi-Epitope Vaccine for Q Fever in Animal Models of Coxiella burnetii Immunity. Frontiers in Immunology. 2022 May 16;13:901372.
76. Miller HK, Kersh GJ. Analysis of recombinant proteins for Q fever diagnostics. Scientific Reports. 2020 Dec 1;10(1):20934.
77. Hogerwerf L, van den Brom R, Roest HI, Bouma A, Vellema P, Pieterse M, et al. Reduction of Coxiella burnetii prevalence by vaccination of goats and sheep, The Netherlands. Emerging Infectious Diseases. 2011 Mar;17(3):379-56.
78. Marmion BP, Ormsbee RA, Kyrkou M, Wright J, Worswick DA, Izzo AA, et al. Vaccine prophylaxis of abattoir-associated Q fever: eight years' experience in Australian abattoirs. Epidemiology & Infection. 1990 Apr;104(2):275-87.
79. Long CM. Q fever vaccine development: current strategies and future considerations. Pathogens. 2021 Sep 22;10(10):1223.
80. Sam G, Stenos J, Graves SR, Rehm BH. Q fever immunology: the quest for a safe and effective vaccine. NPJ Vaccines. 2023 Sep 7;8(1):133.
81. Chen C, Dow C, Wang P, Sidney J, Read A, Harmsen A, et al. Identification of CD4+ T cell epitopes in C. burnetii antigens targeted by antibody responses. PLoS One. 2011 Mar 15;6(3):e17712.
82. Xiong X, Qi Y, Jiao J, Gong W, Duan C, Wen B. Exploratory study on Th1 epitope-induced protective immunity against Coxiella burnetii infection. PLoS One. 2014 Jan 30;9(1):e87206.
83. Lu S, Zgurskaya HI. MacA, a periplasmic membrane fusion protein of the macrolide transporter MacAB-TolC, binds lipopolysaccharide core specifically and with high affinity. Journal of Bacteriology. 2013 Nov 1;195(21):4865-72.
84. Parker CT, Russell R, Njoroge JW, Jimenez AG, Taussig R, Sperandio V. Genetic and mechanistic analyses of the periplasmic domain of the enterohemorrhagic Escherichia coli QseC histidine sensor kinase. Journal of Bacteriology. 2017 Apr 15;199(8):10-128.
85. Xie W, Dickson C, Kwiatkowski W, Choe S. Structure of the Cytoplasmic Segment of Histidine Kinase Receptor QseC: A Key Player in Bacterial Virulence. Protein and Peptide Letters. 2010 Nov 1;17(11):1383-91.
86. Xu Y, Sim SH, Song S, Piao S, Kim HM, Jin XL, et al. The tip region of the MacA α-hairpin is important for the binding to TolC to the Escherichia coli MacAB–TolC pump. Biochemical and Biophysical Research Communications. 2010 Apr 16;394(4):962-5.
87. Sandoz KM, Moore RA, Beare PA, Patel AV, Smith RE, Bern M, et al. β-Barrel proteins tether the outer membrane in many Gram-negative bacteria. Nature microbiology. 2021 Jan;6(1):19-26.