Mini Review
The circular RNA (circRNA) is a covalently closed noncoding RNA, recently with the widespread application of high-throughput RNA sequencing bioinformatics methods, a large number of circRNAs found in human cells have been gradually discovered. It performs multiple biological functions in the human body and participates in the occurrence and development of different diseases such as tumors [1]. Studies have found that circRNA is not easily degraded by exonuclease RNase R, has a half-life of more than 48 hours, can stably exist in eukaryotic cells, and its structure is highly conservative and organized, timing, disease-specific, and is expected to become a potential tumor diagnostic marker and therapeutic target [2].
Multiple myeloma (MM) is a malignant clonal disease that originates from plasma cells and is the second most common malignant tumor of the blood system. It mainly occurs in elderly patients, and multiple data show that the first-line induction, consolidation, and maintenance regimes based on new drugs can significantly improve the overall response rate and overall survival of patients [3-4]. However, the challenges faced by clinicians in treatment such as disease recurrence and multidrug resistance are still common [5-6]. Therefore, in-depth research on mechanisms of drug resistance are urgently needed to improve the long-term prognosis of patients. CircRNA, a special non-coding RNA, is a type of RNA molecule that can be stably expressed and has a highly conserved sequence, which is one of the hot spots in the field of cancer research. This article will illustrate from the discovery and biological function of circRNA, and then discuss the research progress of circRNA in tumor and MM.
The Brief Introduction of CircRNA
In the 1970s, researchers first discovered single-stranded covalently closed RNA in RNA viruses and named it circular RNA (circRNA) [7,8]. Due to the limitation of detection technology, the previously detected circRNA expression levels are extremely low, and has always been considered to be the product of abnormal RNA splicing, which has no actual biological functions. In recent years, with the development of gene sequencing and bioinformatics analysis technology, nearly 100,000 circRNA sequencing results have been collected by Circbase, CircNet, circ2Traits and other databases [9]. Salzman et al. [10] used RNA sequencing technology to detect a large number of circRNA molecules in the bone marrow, Hela cells and human embryonic stem cells (H9) of children with acute lymphoblastic leukemia, confirming that circRNA is widely present in human embryonic stem cells and malignant cells. It has been confirmed that it is a type of RNA that does not have a 5’ -end cap and a 3’ -end poly(A) tail structure. Its unique covalent bond ring structure makes it not affected by RNA exonuclease, and the expression is more stable and not easy to degrade [11]. A number of studies have confirmed that the abnormal expression of circRNA is closely related to tumorigenesis, metastasis, and response to treatment [12].
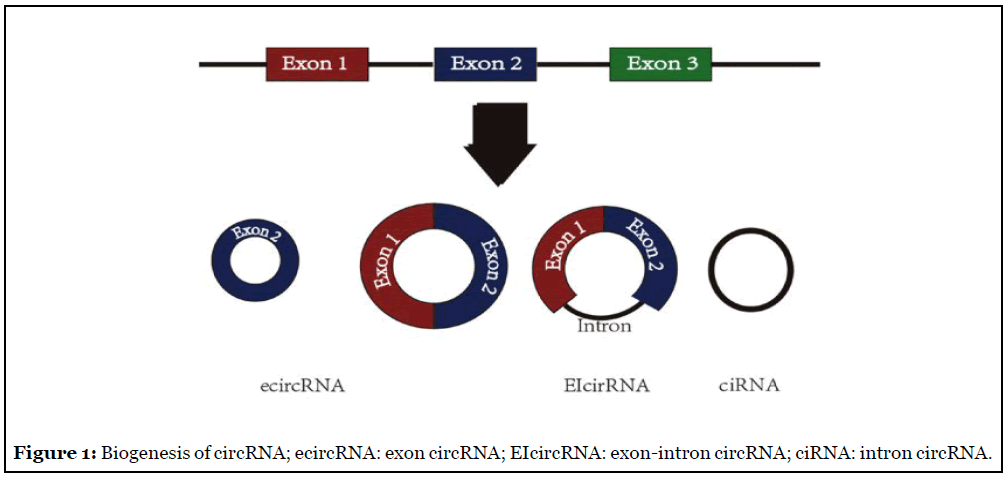
According to the different sequences, circRNA can be divided into three categories [13]: exon circRNA (ecircRNA), circularized intron circRNA (ciRNA) and exon-intron circRNA (EIcircRNA). EcircRNA is composed of exon transcribed RNA and is the most common circRNA, including single exon circRNA and multi-exon circRNA [14], mainly in the cytoplasm. CiRNA is composed of noncoding RNA derived from introns, generally contains 2 or more segments non-coding RNAs derived from introns, mainly present in the nucleus [13,15]. EIcircRNA is composed of two adjacent exons and the intron transcribed between them. It was first discovered in human cells by Li et al. [16] in 2015 and mainly exist in the nucleus. The schematic diagram of circRNA generation is shown in (Figure 1) [17].
The Biological Function of CircRNA
Unlike other non-coding RNAs, the special structure of circRNA makes it having unique biological functions. Researchers have detected abundant and stable circRNA in human exosomes, and circRNA is expected to be a good marker for tumor diagnosis and differential diagnosis [12,18]. CircRNA mainly exerts its biological function through sponge adsorption, cis-regulation of parent gene expression, and participation in protein translation.
Sponge adsorption function
Adsorption of miRNA is the most important function of circRNA. The sponge adsorption function refers to that circRNA can adsorb and release its corresponding miRNA. CircRNA contains a common miRNA reaction element and is a type of competing endogenous RNAs (ceRNAs) [19], which inhibits its biological function by competitively binding miRNAs. There are multiple miRNA binding sites on circRNA, and the ability of circRNA to adsorb miRNA is stronger than other ceRNAs, so it is also called “super sponge”. In 2013, two different research groups, first demonstrated the sponge adsorption function of circRNA at the same time [20,21]. The researchers found that cerebellar degeneration-related protein 1 antisense (CDRlas) were mainly present in human and mouse brain tissues, with a length of about 1500bp. Further research found that ciRS-7/CDRlas and miR-7 had overlapping co-expression. ciRS-7/CDRlas had more than 70 miRNA binding sites. The binding force of CDRlas and miRNA was 10 times that of GADPH. CDRlas inhibits the biological function of miR-7 by binding to miR-7, and up-regulates the expression of miR-7 target genes, such as epidermal growth factor receptor, insulin receptor matrix-1, etc., thereby inhibiting the proliferation, invasion and tumorigenesis of cancer cells. Studies have shown that miR-7 plays a role in promoting cancer in lung cancer [22] and other tumors [23]. As a natural ceRNA, cirRNA is expected to replace anti-nucleotide chemical drugs by regulating miRNA levels to regulate upstream and downstream gene networks and play a role in anti-tumor protection [24].
CircRNA cis-regulates parental gene expression
Another important function of circRNA is to regulate the expression of its parent genes. First, circRNA and RNA binding protein bind to each other to regulate the expression of the parent genes. Second, circRNA is balanced with linear RNA through competitive complementary pairing among introns during its formation, thereby regulating the expression of mRNA and affecting protein translation. CircRNA can also regulate the expression of target genes through the interaction of RNA-DNA, RNA-RNA and RNA-protein to establish an important biological network. The study of Lai et al. [25] proved that hsa_circ_0047905 and hsa_circ_0138960 were positively correlated with the expression levels of their parental genes SERPINB5 and GDAmRNA. After knocking down hsa_circ_0047905 and hsa_circ_0138960 in gastric cancer cells, the parental gene expression level decreased significantly. CircRNA ciankrd52 is mainly located in the nucleus and enriched in its transcribed region. Ci-ankrd52 regulates the expression of the parental gene by regulating the prolonged activity of RNA polymerase II and such circRNA located in the nucleus can regulate the expression of the parental gene [26]. LiF et al. [16] found that in esophageal cancer, circ- ITCH can regulate the expression of its parental gene ITCH by interacting with miR-7, miR-17 and miR-214.
Role of cirRNA in protein translation
Although circRNA is a non-coding RNA, studies have shown that a small number of circRNAs have the function of translating proteins. The function of circRNA-encoding proteins was first confirmed in hepatitis D virus, can encode protein with 122 amino acids [27]. Wang et al. [28] through constructing a microgene containing cytomegalovirus promoter, internal ribosome entry site (IRES) and exon encoding green fluorescent protein (GFP), transferred it into the cell to form circRNA, which can be translated into GFP protein. Legnini et al. [29] further found that circ- ZNF609 has an open reading frame, with a start codon and a stop codon at both ends, which are translated into protein by polysome translation.
Pamudurti et al. [30] applied mass spectrometry and found that circZNF609 from the myogenic site can encode a protein that is involved in the process of muscle development. Yang et al. [31] also found that N6-methyladenine promotes the initiation of circRNA translation. Zhang et al. [32] found that the second exon of linc-PINT can form circ-PINT by self-cyclization, and confirmed that circPINT drives translation through an IRES of a new peptide consisting of 87 amino acids PINT87aa, which can inhibit the occurrence and development of malignant glioma.
Other functions
Circ-Foxo3 can combine with cycle-related proteins CDK2 and p21 to form RNA-protein complexes, thereby inhibiting the relationship between CDK2 protein and cyclin A and E, and inhibiting cell cycle progression [33]. CircHIPK2 targets miR-124-2HG at the post-transcription level in combination with autophagy and endoplasmic reticulum stress to regulate astrocyte activation [34].
CircRNA and Tumor
CircRNA is closely related to the occurrence and development of tumors, and plays an important role in the diagnosis, differential diagnosis and prognosis of tumors. Specific circRNAs can be detected in saliva, exosomes and clinical blood samples, which will make them reliable biomarkers for potential diagnosis or prediction of tumor efficacy and even therapeutic targets. With the development of second-generation sequencing technology, more than 1 million circRNAs have been detected in human tissues, mostly in the cytoplasm, and their abundance far exceeds that of homologous linear RNA. It has been reported that circRNA is closely related to the occurrence and development of gastric cancer, colon cancer, breast cancer, lung cancer, liver cancer, head and neck tumor, cervical cancer, hematological malignancies and other malignant conditions [12,16,17,25,27,29,35].
Dahl M et al. [35] used NanoString technique to perform genome-wide gene sequencing on four different mantle cell lymphoma cell lines REC-1, Granta 519, UPN2, Z138 and MM cell lines NTCI-H929, and detected numerous cirRNAs, CIRS-7, circHIPK3, circCCDC66, circFBXW7, circSMARCA5, circCDYL and circZKSCAN1, while these cirRNA also showed different levels of expression in other tumors. In addition, circRNAs associated important genes involved in lymphoma and MM were also detected, including FOXP1, SETD3, EZH2, ATM, XPO1, IKZF3, CD11Aand WHSC1. Among them, circRNAIKZF3 from IKZF3 gene was highly expressed in MM cell line NCI-H929, which is not listed in circBase and has, to our knowledge, not previously been reported [9]. Based on this, cirRNA may be a candidate indicator to distinguish different B-cell malignancies in the future. Circ-RAD23B is an oncogene in non-small cell lung cancer, and high expression of circ- 0001017 and circ-0061276 are associated with better prognosis in gastric cancer [37]. The expression level of circ-0004277 in patients with acute myeloid leukemia was significantly lower than that in the normal population, and the expression level of circ-0004277 increased significantly after successful chemotherapy. In acute myeloid leukemia it has a certain diagnostic role and has a predictive therapeutic effect [38]. Sui et al. [39] analyzed circRNA and mRNA in gastric cancer tissues and adjacent tissues, and found that as many as 69 circRNAs were significantly differentially expressed, which were mainly absorbed by the sponge to regulate the expression level of target genes and play their biological functions. circRNAmiRNA and miRNA-mRNA directly/indirectly promote the biological progress of gastric cancer by regulating the expression of CD44, CXXC5, MYH9, MALAT1 and other genes. Xia et al. [40] have pointed out that circ-CBFB (hsa_ circ_0000707) is significantly higher expressed in chronic lymphocytic leukemia (CLL) than in the normal control group. Circ-CBFB overexpression is an independent prognostic factor for CLL. As a sponge factor of mir-607, circ-CBFB targets FZD3 downstream and promotes tumor development by targeting the Wnt/β-catenin signaling pathway. Therefore, circ-CBFB-miR-607-FZD3-Wnt/β-catenin axis may be a potential therapeutic target for CLL.
F-circRNA is derived from chromosomal heterotopic or fusion circRNA generated by gene fusion. Such f-circrNA is involved in tumor progression and may serve as a further diagnostic and therapeutic target. F-circRNA of PML-RAR and MLL-AF9 have been found in hematopoietic tumor [41]. Pan et al. [42] reported that circ-BA9.3 is an f-CircRNA with 1,137 nucleotides and is a source of BCR-ABL1, and circ-BA9.3 is involved in the resistance of tyrosine kinase inhibitor. In patients with tyrosine kinase inhibitor resistance, circ-BA9.3 was significantly increased and positively correlated with BCR-ABL1 expression. Circ-BA9.3 was also found to improve the translation efficiency of BCR-ABL1 or inhibit the degradation of oncoproteins. Circ-BA9.3 may enhance resistance to tyrosine kinase inhibitor drugs such as imatinib, dasatinib and nilotinib by increasing the protein level of BCR-ABL1 oncogene.
CircRNA and Multiple Myeloma
Patients with MM eventually relapse and develop drug resistance, which forces us not to ignore the basic research on MM, but still need to find new diagnosis and treatment, prognosis assessment, and new targets to overcome drug resistance. The research of circRNA is still in its infancy, and its mechanism and biological function still need to be clarified. Moreover, studies on circRNA in MM are rare, and more in-depth studies are needed to clarify its role and possible mechanisms in MM.
CircRNA diagnostic role in MM
Our research group has detected the bone marrow samples of 4 MM patients and 4 normal subjects by circRNA expression profile chip technology. A total of 122 up-regulated circRNAs and 260 down-regulated circRNAs were screened out, and the weighted ranking of circRNAs according to Log2FC showed that circRNA PTK2 was significantly highly expressed in MM, with the most significant difference between MM and the normal control group, which could be used as a good indicator of MM diagnosis [43]. Xiao Liu et al. [44] showed that hsa_circRNA_101237 was highly expressed in MM patients and could be used as a diagnostic marker. Meng Gao et al. results showed that hsa_circ_0007841 was highly expressed in drug-resistant myeloma cell lines and myeloma patients, and the area under ROC curve was 0.907, playing a diagnostic role [45].
Relationship between circRNA and MM treatment and prognosis
The expression of hsa_circRNA_101237 was significantly higher in MM patients, and was especially increased in 13q14 deletion, 1q21 amplification, P53 deletion, t(4,14) and t(14,16) patients [44], and these patients had a poor prognosis [46]. Studies have shown that hsa_ circRNA_101237 was significantly associated with poor OS and PFS [44]. CircRNA plays an important role in the regulation of MM cell resistance. CircRNA0007841 was highly expressed in drug-resistant MM cell lines [45]. In MM, circRNA0007841 was significantly correlated with MM type, cytogenetics, bone destruction, and R-ISS stage, and its high expression was significantly correlated with 1q21 amplification, t (4:14), ATR, and IRF4 gene mutations, with poor prognosis. The bioinformatics results showed that has_circ_0007841 interacted with 8 miRNA and 10 mRNA, among which has_circ_0007841 promoted tumor progression by sponge adsorption of hsa-miR- 29b-2-5p [45]. Because miR29b could inhibit osteoclast differentiation, thus reversing the osteoclast activation caused by myeloma and delaying myeloma progression. In addition, miR29b can promote bortezomib induced apoptosis of myeloma cells [47], so MM patient with high expression of has_circ_0007841 has a poor prognosis.
CircRNA was used as competitive endogenous circRNA (ceRNA), and circUBAP2 was competitive in inhibiting miR-143 and blocking MM cell apoptosis. Bcl-2 family is a key molecule in the apoptosis pathway, and one of the targets of miR-143 is bcl-2, so it is involved in the apoptosis pathway of MM cells [48,49]. Liu et al. [50] found in 105 MM patients and 36 normal controls subjects that circ- SMARCA5 was negatively correlated with ISS stage and β2 microglobulin, and its expression was correlated with complete response rate and better PFS and OS. Circsmarca5 inhibits proliferation and induces apoptosis of myeloma cells by targeting miR-767-5p.
In our verification analysis of 60 MM patients and 30 normal subjects, RT-PCR results showed that the expression level of circ-PTK2 in bone marrow of MM patients was significantly higher than that of normal subjects, and the PFS of patients in the high-expression group was significantly lower than that in the lowexpression group [43]. High circRNA PTK2 expression is associated with a poorer treatment response and shorter prognostic survival. Circ-PTK2 may inhibit apoptosis by regulating the proliferation and invasion of MM cells, leading to poor prognosis [43]. Chen et al. [51] showed that circ-CDYL, as a sponge molecule of miR-1180, promoted the expression of YAP protein and promoted the progress of MM.
Feng et al. [52] confirmed that circ-0000190 inhibits MM progression by targeting miR-767-5p/MAPK4 signaling pathway. The overexpression of circ-0000190 significantly reduced the activity and proliferation of myeloma cells and inhibited the cell cycle in G1 phase by down-regulating CDK6, CDK4, CyclinD1, CyclinEI and up-regulating p21Clip1.
The circRNA screened in the current study mostly play the role of oncogenes [39,40,44,45,53] and inhibit the proliferation and invasion of tumor cells by silencing the circRNA and/or its mediated signaling pathway, and the inhibition of apoptosis is becoming a new hotspot in cancer therapy [53-56]. One study showed that silencing RNA-ZNF652 could increase the expression of miR-205, thus affecting the process of epithelial-mesenchymal transformation, thereby inhibiting the proliferation of renal cell carcinoma and promoting apoptosis [53]. Circ- CDYL knockdown significantly blocked the growth of MM cells [51]. With the development of gene editing, FDA granted CRISPR/Cas9 gene-edited hematopoietic stem cell therapy CTX001 as a fast-track treatment for transfusiondependent β-Thalassaemia. The clinical application of circRNA is just around the corner.
Great progress has been made in the study of circRNA in tumor development, invasion and metastasis, but there are still many problems to be further explored. First, the naming system of circRNA has not been unified yet. The role of circRNA in nuclear and cytoplasmic transport needs to be further clarified. Second, the mechanism of post-transcriptional regulation of circRNA remains to be explored. For example, it is not known how they are eventually degraded and how their structures differ in function from their linear counterparts. Third, in addition to the classical reverse splicing and exon jumping to form circRNA, other patterns of splicing or post-transcriptional modification may exist, such as template switching and tandem replication by reverse transcriptase [57].
At present, the research of circRNA is still in the basic research stage, and there are still some deficiencies in clinical application. For example, the application of circRNA in molecular markers of disease is still lacking in large sample clinical verification, and it is difficult to achieve real clinical transformation at present. The present study is more focused on circRNA sponges adsorption function by studying the endogenous cyclic sponge structure, binding sites, design and development of an effective artificial sponge. According to its special formation mechanism, new drug research can enhance good circRNA expression and silent bad circRNA in the human body without side effects by regulating circRNA abundance.
It is believed that with the improvement of highthroughput sequencing technology and the reduction of cost, as well as the continuous improvement of calculation methods, more circRNA molecules will be predicted accurately, laying a foundation for the exploration of biological markers of MM and subsequent functional studies. Its exploration in MM will also be further expanded which will eventually leads to clinical benefit of patients.
References
2. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013 Feb 1;19(2):141- 157.
3. Bianchi G, Richardson PG, Anderson KC. Promising therapies in multiple myeloma. Blood, The Journal of the American Society of Hematology. 2015 Jul 16;126(3):300- 310.
4. Kortüm KM, Einsele H. Diagnostic and therapeutic considerations on recurrence of multiple myeloma: A current overview. Der Internist. 2019 Jan 1;60(1):34-41.
5. Caillon H, Avet-Loiseau H, Attal M, Moreau P, Decaux O, Dejoie T. Comparison of sebia free light chain assay with freelite assay for the clinical management of diagnosis, response, and relapse assessment in multiple myeloma. Clinical Lymphoma Myeloma and Leukemia. 2019 May 1;19(5):e228-237.
6. Pinto V, Bergantim R, Caires HR, Seca H, Guimarães JE, Vasconcelos MH. Multiple Myeloma: Available Therapies and Causes of Drug Resistance. Cancers. 2020 Feb;12(2):E407.
7. Hewlett MJ, Pettersson RF, Baltimore D. Circular forms of Uukuniemi virion RNA: an electron microscopic study. Journal of Virology. 1977 Mar 1;21(3):1085-1093.
8. Diener TO. Potato spindle tuber “virus”: IV. A replicating, low molecular weight RNA. Virology. 1971 Aug 1;45(2):411-428.
9. Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clinical Chemistry. 2015 Jan 1;61(1):221-230.
10. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One. 2012 Feb 1;7(2):e30733.
11. Chen LL. The biogenesis and emerging roles of circular RNAs. Nature Reviews Molecular Cell Biology. 2016 Apr;17(4):205-211.
12. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018 Feb;37(5):555-565.
13. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Molecular Cancer. 2017 Dec 1;16(1):58.
14. Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. International Journal of Molecular Sciences. 2014 Jun;15(6):9331-9342.
15. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Molecular Cell. 2013 Sep 26;51(6):792-806.
16. Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/ß-catenin pathway. Oncotarget. 2015 Mar 20;6(8):6001-6013.
17. Mei M, Wang Y, Li Z, Zhang M. Role of circular RNA in hematological malignancies. Oncology Letters. 2019 Nov 1;18(5):4385-4392.
18. Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. Journal of hematology & oncology. 2018 Dec 1;11(1):21.
19. Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Molecular Cancer. 2018 Dec;17(1):79.
20. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013 Mar;495(7441):333-338.
21. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013 Mar;495(7441):384-388.
22. Chou YT, Lin HH, Lien YC, Wang YH, Hong CF, Kao YR, et al. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Research. 2010 Nov 1;70(21):8822-8831.
23. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Research. 2013 Sep 15;73(18):5609-5612.
24. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014 Jan;505(7483):344-352.
25. Lai Z, Yang Y, Yan Y, Li T, Li Y, Wang Z, et al. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_ circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle. 2017 Dec 2;16(23):2301-2311.
26. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Molecular Cell. 2013 Sep 26;51(6):792-806.
27. He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Letters. 2017 Jun 28;396:138-144.
28. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015 Feb 1;21(2):172- 179.
29. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Molecular Cell. 2017 Apr 6;66(1):22-37.
30. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of circRNAs. Molecular Cell. 2017 Apr 6;66(1):9-21.
31. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N 6-methyladenosine. Cell Research. 2017 May;27(5):626- 641.
32. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nature Communications. 2018 Oct 26;9(1):4475.
33. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Research. 2016 Apr 7;44(6):2846-2858.
34. Huang R, Zhang Y, Han B, Bai Y, Zhou R, Gan G, et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124–2HG. Autophagy. 2017 Oct 3;13(10):1722-1741.
35. Dahl M, Daugaard I, Andersen MS, Hansen TB, Grønbæk K, Kjems J, et al. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Laboratory Investigation. 2018 Dec;98(12):1657-1669.
36. Han W, Wang L, Zhang L, Wang Y, Li Y. Circular RNA circ-RAD23B promotes cell growth and invasion by miR- 593–3p/CCND2 and miR-653–5p/TIAM1 pathways in non-small cell lung cancer. Biochemical and Biophysical Research Communications. 2019 Mar 12;510(3):462-466.
37. Li T, Shao Y, Fu L, Xie YI, Zhu L, Sun W, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. Journal of Molecular Medicine. 2018 Jan 1;96(1):85-96.
38. Li W, Zhong C, Jiao J, Li P, Cui B, Ji C, et al. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. International Journal of Molecular Sciences. 2017 Mar;18(3):597.
39. Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J, et al. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncology Reports. 2017 Feb 28;37(3):1804-1814.
40. Xia L, Wu L, Bao J, Li Q, Chen X, Xia H, et al. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/ß-catenin pathway. Biochemical and Biophysical Research Communications. 2018 Sep 3;503(1):385-390.
41. Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016 Apr 7;165(2):289- 302.
42. Pan Y, Lou J, Wang H, An N, Chen H, Zhang Q, et al. CircBA9. 3 supports the survival of leukaemic cells by up-regulating c-ABL1 or BCR-ABL1 protein levels. Blood Cells, Molecules, and Diseases. 2018 Nov 1;73:38-44.
43. Zhou F, Wang D, Wei W, Chen H, Shi H, Zhou N, et al. Comprehensive profiling of circular RNA expressions reveals potential diagnostic and prognostic biomarkers in multiple myeloma. BMC Cancer. 2020 Dec;20(1):40.
44. Liu X, Tang H, Liu J, Wang X. hsa_circRNA_101237: A Novel Diagnostic and Prognostic Biomarker and Potential Therapeutic Target for Multiple Myeloma. Cancer Management and Research. 2020 Mar 20;12:2109- 2118.
45. Gao M, Xiao H, Hang D, Jiang S, Fu Y, Gong L. hsa_circ_0007841: a novel potential biomarker and drug resistance for multiple myeloma. Frontiers in Oncology. 2019 Nov 19;9:1261.
46. Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NB, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015 Nov 20;33(33):3911.
47. Amodio N, Di Martino MT, Foresta U, Leone E, Lionetti M, Leotta M, et al. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death & Disease. 2012 Nov; 3 (11): e436.
48. Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Molecular cancer. 2018 Dec;17(1):79.
49. Zhong Y, Du Zhang H, Wang G, Ding C, Liu P, Wang R, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017 Sep 5;8(37):61687- 61697.
50. Liu H, Wu Y, Wang S, Jiang J, Zhang C, Jiang Y, et al. Circ-SMARCA5 suppresses progression of multiple myeloma by targeting miR-767-5p. BMC Cancer. 2019 Dec 1;19(1):937.
51. Chen F, Wang X, Fu S, Wang S, Fu Y, Zhang J, et al. Circular RNA circ-CDYL sponges miR-1180 to elevate yesassociated protein in multiple myeloma. Experimental Biology and Medicine. 2020 Jun;245(11):925-932.
52. Feng Y, Zhang L, Wu J, Khadka B, Fang Z, Gu J, et al. CircRNA circ_0000190 inhibits the progression of multiple myeloma through modulating miR-767-5p/ MAPK4 pathway. Journal of Experimental & Clinical Cancer Research. 2019 Dec;38(1):54.
53. Zhang L, Guo Y. Silencing circular RNA-ZNF652 represses proliferation and EMT process of renal carcinoma cells via raising miR-205. Artificial Cells, Nanomedicine, and Biotechnology. 2020 Jan 1;48(1):648-655.
54. Liu T, Ye P, Ye Y, Lu S, Han B. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. Journal of Cellular and Molecular Medicine. 2020 Feb;24(3):2189-2201.
55. Chi G, Yang F, Xu D, Liu W. Silencing hsa_circ_ PVT1 (circPVT1) suppresses the growth and metastasis of glioblastoma multiforme cells by up-regulation of miR-199a-5p. Artificial Cells, Nanomedicine, and Biotechnology. 2020 Jan 1;48(1):188-196.
56. Yang D, Qian H, Fang Z, Xu A, Zhao S, Liu B, et al. Silencing circular RNA VANGL1 inhibits progression of bladder cancer by regulating miR-1184/IGFBP2 axis. Cancer Medicine. 2020 Jan;9(2):700-710.
57. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Molecular Cell. 2018 Aug 2;71(3):428-442.
