Keywords
Cancer, Metabolism, Mitochondria, Oxidative stress, Omics
Metabolism Reprogramming and Redox Signaling in Cancer
Although being a complex pathology with genetic as well as non-genetic etiologies, cancer is characterized by a limited series of hallmarks including energetic metabolism reprogramming [1]. Long ago O. Warburg unraveled a bias of tumor metabolism in favor of glycolysis, the socalled Warburg effect. Indeed, tumor cells display both an increase in glucose uptake and fermentation of glucose to lactate [2]. Since these pioneering works, different alterations of tumor metabolism were reported such as deregulated uptake of glutamine or new modes of nutrient acquisition [3]. In general, cancer cells exhibit a high rate of proliferation due to constitutive anabolism and catabolism activation. Moreover, by competing for nutrients, inducing hypoxia and releasing inhibitory molecules in tumor microenvironment (TME), tumors dramatically affect immune cell metabolism [4].
Cellular production of ATP relies on both glycolysis and oxidative phosphorylation (OXPHOS) also called mitochondrial respiration. In cancer cells, the reprogramming of ATP production from OXPHOS to glycolysis manifests by major consequences including enhanced glucose consumption, aberrant cell death signaling, enhanced oxidative stress and autophagy. This oxidative stress challenges redox homeostasis through (i) down regulation of antioxidant enzymes and (ii) activation of processes contributing to reactive oxygen species (ROS) production, which will modify cellular responses and cell fate [5]. For example, at the plasma membrane level, pro-oxidant enzymes such as NADPH oxidase (NOX) contributes to ROS production whereas intracellularly, the mitochondrial electron transport chain constitutes a major source of anion superoxide (O2.-) that is rapidly converted to hydrogen peroxide (H202). Antioxidant enzymes (e.g., superoxide dismutase, catalase, thioredoxin, glutathione reductase) are also frequently deregulated in tumors leading to changes in ratio of the redox buffers, GSSG/GSH/ GSSG, NADH/NAD+ and NADPH/NADP+. Consequently, this affects post-translational modification of proteins and their function influencing many basic biochemical processes and favoring tumorigenesis, metastasis and drug resistance [6,7].
Establishing Cancer Energy Metabolic Profile
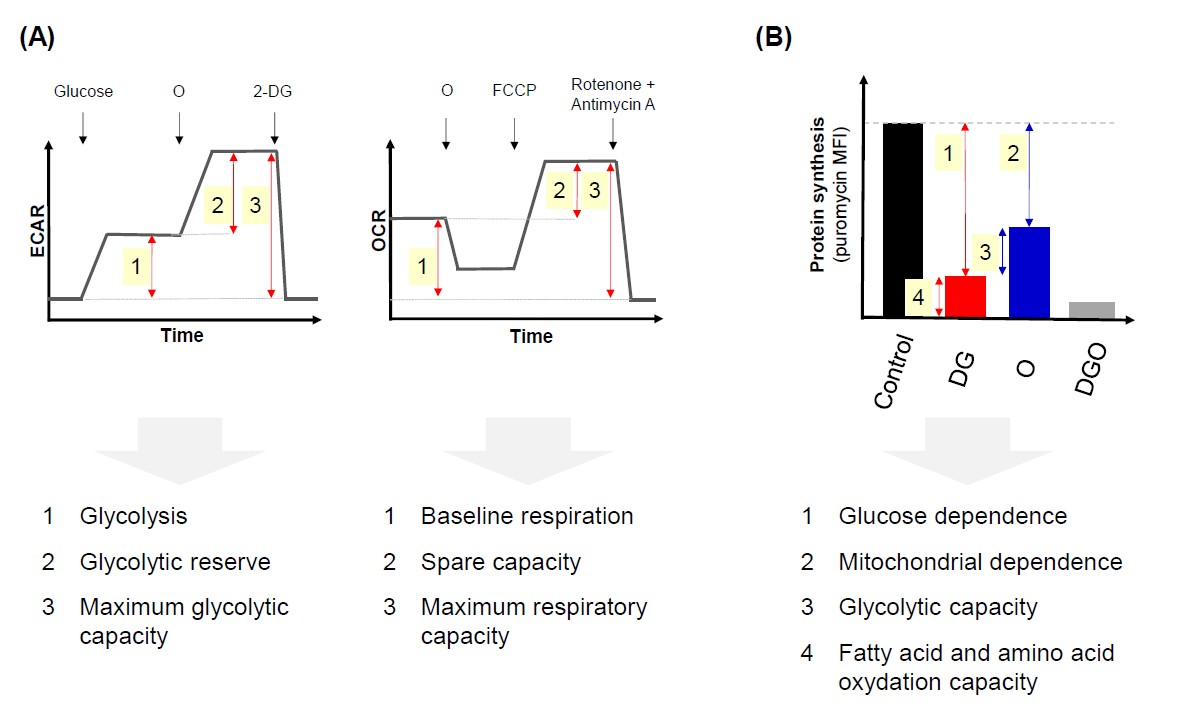
Metabolic alterations of cancer cells are generally evaluated by bulk methods including (i) Seahorse technology (ii) maximal enzymatic activities and (iii) mass spectrometry. The Seahorse technology (Agilent) relies on the monitoring of two parameters in tumor cell extracellular flux: extracellular acidification rate (ECAR) and oxygen consumption rate (OCR), corresponding to glycolysis and OXPHOS, respectively, using fluorescence detection in microtiter plates. The measurement of ECAR allows to determine glycolysis, glycolytic reserve and maximum glycolytic capacity while OCR helps to discriminate between baseline, sparse and maximum mitochondrial respiration (Figure 1A). This Seahorse technology is adapted to tumor cells in culture. However, it is confronted to some drawbacks such as the requirement of a large number of cells (e.g. from 104 to 106 cells/well in 96-well plate) and the requirement for cell culture (about several hours to one day) that could alter cell metabolism. Besides Seahorse technology, tumor cell metabolism could be measured by monitoring maximal enzymatic activities such as lactate dehydrogenase activity using cell culture supernatants or tumor cell lysates. Also, mass spectrometry is used to monitor the levels of metabolites (hundred to thousands) produced by different metabolic pathways. As tumors are composed of mixed cell populations and cell metabolism exhibits an intrinsic plasticity, a crucial challenge was to develop quantitative methods to profile metabolism and its potential alterations at the single cell level. This challenge has now been overcome through the development of SCENITH (single-cell energetic metabolism by profiling translation inhibition).
(B) SCENITH technology monitors the level of protein synthesis as a surrogate of ATP production. Puromycin antibiotic able to be incorporated in new synthesized proteins is used as a means to monitor protein synthesis. Briefly, cells are mock treated (control) or exposed for a short time with DG, O or both and puromycin is added in the last minutes of incubation. Then after intracellular staining with a monoclonal antibody specific for puromycin, the mean fluorescence intensity (MFI) corresponding to the efficiency of puromycin incorporation is determined in each condition by cell cytometry. The comparison of MFI between untreated cells and cells treated with metabolic inhibitors leads to determination of 4 parameters: (i) glucose dependence, (ii) mitochondrial dependence, (iii) glycolytic capacity and (iv) fatty acid and amino acid oxidation capacity. Adapted from (Argüello et al. [8]).
Introducing SCENITH
SCENITH method is based on the analysis of protein synthesis (PS) by flow cytometry [8]. This method relies on the fact that about 50% of ATP production is consumed by the PS process. Therefore, Arguëllo et al. proposed to monitor PS as a surrogate of ATP production. Briefly, SCENITH is based on the use of puromycin, an antibiotic whose incorporation in nascent polypeptide chain is followed after cell permeabilization using a specific monoclonal antibody as a read out. In addition, this flow cytometry method combines elegantly glycolysis inhibition with 2-deoxy-D-glucose (2-DG) and OXPHOS inhibition with oligomycin (O) to determine energetic metabolism capacities and substrate dependencies to glucose, amino acids and fatty acids (Figure 1B). First using different inhibitors Argüello et al. provide evidence that puromycin incorporation correlates with ATP production. Then they applied this observation to the establishment of energetic metabolism profile. In order to validate this new method Argüell et al. compare its ability to establish the energy metabolic profile of T cells to the Seahorse technology. More precisely, peripheral blood T cells from healthy donors were unstimulated or stimulated with anti-CD3/ CD28 antibodies and their metabolic profile was compared by the two methods. As demonstrated previously by others, upon stimulation T-cells shifted to aerobic glycolysis with comparable results with SCENITH and Seahorse methods. Interestingly, SCENITH requires 10-fold fewer cells than Seahorse and allow to evaluate in the same time T-cell phenotypes.
To further demonstrate the potency of SCENITH, Argüello et al. applied this technology to whole blood and human tumor samples ex vivo. First, they showed that this technique was able to reveal that early effector memory (EEM) CD8+ T cells from human blood present a high glycolytic capacity. Such a feature was not found using Seahorse method because the signal from EEM CD8+ population was masked by the metabolism of other blood cells. Also, Argüllelo et al. investigated the metabolic profile of dendritic cell (DC) subsets before and after activation and revealed that upon activation by Toll-like receptor agonists type 1 and type 2 DC shifted their metabolism to lower mitochondrial dependence. Finally, they studied the metabolism profile of myeloid cells purified from tumors showing for example strong mitochondrial dependence of tumor associated macrophages (TAM). Interestingly, they combined SCENITH and single-cell RNA-seq analysis of myeloid populations purified from renal carcinomas and juxta-tumoral tissues and succeeded to correlate metabolic profiling with metabolic gene expression. Thus, SCENITH is a simple and sensitive new method to evaluate metabolic capacities and dependencies via metabolic profiling of small immune cell populations derived from fresh blood as well as solid tumors [8].
Characterization of γδ T-cell Metabolism by SCENITH
As stated above, tumor metabolism and in particular high tumor glucose consumption has a strong impact on immune cells in the tumor microenvironment [4]. Several studies have investigated this field by analyzing the metabolism of different immune cell types. Very recently the power of SCENITH method was applied to better understand the biology of tumor-infiltrating γδ T-cells. While interferon γ (IFNγ)-producing γδ T cells are associated with tumor control, interleukin-17 (IL- 17) expressing γδ T cells are linked to tumor growth [9]. Because of the scarcity of both populations in TME, their metabolic profile could not be assessed using Seahorse technology. However, using SCENITH, Lopes et al. were able to show that IFNγ-producing γδ T cells display a glycolytic profile while IL-17-producing γδ T cells relies on mitochondrial activity and lipid oxidation [10]. They also found that these metabolic profiles were established during γδ T-cell thymic development. Interestingly, Myc a factor controlling the transcription of glycolytic enzymes was shown to be upregulated in IFNγ-producing γδ T cells. These observations pushed Lopes et al. to propose to implement glucose to favor the development of IFNγ- producing γδ T cells in ex vivo protocols developed for anti-tumor adoptive cell therapy [10].
Adapting SCENITH to Redox Signaling Analysis
Since SCENITH is based on the use of metabolic inhibitors, namely O, rotenone, one can imagine using other classes of chemical inhibitors modulating metabolism on a variety of different cell types. For example, use of ROS scavengers or antioxidants such as N-acetylcysteine, vitamin E or tempol could help to decipher whether oxidative stress influence metabolic profile at the single cell level. Similarly, the use of inhibitors targeting the glutathione metabolism could inform specifically on the implication of this process in energetic metabolism. This could clearly help to evaluate the contribution of redox signaling to tumoral metabolism but also to a myriad of pathologies involving metabolism deregulation such as obesity, metabolic syndrome, celiac disease as well as diabetes. Calcium signaling knowledge could also largely benefit from SCENITH using calcium chelators or proteins inhibitors targeting intracellular calcium stores such as the endoplasmic reticulum, the nucleus and mitochondria. Moreover, SCENITH might have tremendous implications in other physio-pathological conditions. It could also help to have a better knowledge of cell death pathways, which have putative multiple connections with metabolism and redox imbalance as revealed recently in ferroptosis [11]. In another context, this approach could promote a better understanding of cellular metabolic changes in organs affected by chronic pathologies. This could apply to the liver that is submitted to inflammation, lipid overload stress in non-alcoholic steatohepatitis (NASH) or viral infection [12]. The liver being composed of various cell types (i.e. hepatocytes, immune cells, fibroblasts, stellate cells) and under the systemic influence of fat tissue [13], SCENITH could help to develop new insights and a better integrated view of the metabolism contribution to liver pathologies.
Limitations and Open Questions
Although the SCENITH technique holds a lot of promise, it seems to be dedicated to the analysis of subpopulations rather than strictly to the analysis of single cells. Indeed, the study of metabolism is conducted by evaluating 4 experimental conditions in parallel and not on the same cell. Also, since SCENITH relies on the measurement of the mean fluorescence intensity (MFI) of puromycin, at least several hundred of cells should be analyzed for each conditions. Moreover, SCENITH interest would be limited in cell types that do not have an important level of PS such as mature spermatozoids or quiescent stem cells. Thus, it would be interesting for the scientific community to build an exhaustive catalog of cell types that are eligible to SCENITH analysis. In addition, the PS process has its proper temporal kinetic of modulation. Then, it is plausible that cell metabolism in acute or chronic stresses would be difficult to investigate by SCENITH because on their impact on PS. Interestingly, Argüello et al. proposes that their method could be combined with the detection of phenotypic markers, fluorescent lipids or mitochondrial tracers. However, each protocol modification will require a rigorous optimization and standardization notably at the level of cell isolation in order to limit metabolic effects. Finally, the future will tell us whether this technique can be implemented to generate major advances in basic science and could be transferred from bench to bedside and help for precision medicine improvement.
Acknowledgments
K. Benihoud wish to thank CNRS (Centre National de la Recherche Scientifique) and ANR (agence nationale de la recherche) for their financial support. C. Brenner would like to thank CNRS, University Paris-Saclay, Gustave Roussy Institute, INCA (Institut national du Cancer) and SFCE (Société Francaise des Cancers de l’Enfant) for funding her research and METSY laboratory.
References
2. Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nature Reviews Cancer. 2011 May;11(5):325-37.
3. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metabolism. 2016 Jan 12;23(1):27-47.
4. Guerra L, Bonetti L, Brenner D. Metabolic modulation of immunity: a new concept in cancer immunotherapy. Cell Reports. 2020 Jul 7;32(1):107848.
5. Clément MV, Pervaiz S. Intracellular superoxide and hydrogen peroxide concentrations: a critical balance that determines survival or death. Redox Report. 2001 Aug 1; 6(4):211-4.
6. Cai Z, Yan LJ. Protein oxidative modifications: beneficial roles in disease and health. Journal of Biochemical and Pharmacological Research. 2013 Mar;1(1):15-26.
7. Marengo B, Nitti M, Furfaro AL, Colla R, Ciucis CD, Marinari UM, Pronzato MA, Traverso N, Domenicotti C. Redox homeostasis and cellular antioxidant systems: crucial players in cancer growth and therapy. Oxidative Medicine and Cellular Longevity. 2016 Oct; 2016.
8. Argüello RJ, Combes AJ, Char R, Gigan JP, Baaziz AI, Bousiquot E, et al. SCENITH: a flow cytometrybased method to functionally profile energy metabolism with single-cell resolution. Cell Metabolism. 2020 Dec 1;32(6):1063-75.
9. Lopes N, Silva-Santos B. Functional and metabolic dichotomy of murine γδ T cell subsets in cancer immunity. European Journal of Immunology. 2021 Jan;51(1):17-26.
10. Lopes N, McIntyre C, Martin S, Raverdeau M, Sumaria N, Kohlgruber AC, et al. Distinct metabolic programs established in the thymus control effector functions of γδ T cell subsets in tumor microenvironments. Nature Immunology. 2021 Feb; 22(2):179-92.
11. Weiland A, Wang Y, Wu W, Lan X, Han X, Li Q, Wang J. Ferroptosis and its role in diverse brain diseases. Molecular Neurobiology. 2019 Jul; 56(7):4880-93.
12. Brenner C, Galluzzi L, Kepp O, Kroemer G. Decoding cell death signals in liver inflammation. Journal of Hepatology. 2013 Sep 1;59(3):583-94.
13. Ingaramo MC, Sánchez JA, Perrimon N, Dekanty A. Fat Body p53 Regulates Systemic Insulin Signaling and Autophagy under Nutrient Stress via Drosophila Upd2 Repression. Cell Reports. 2020 Oct 27;33(4):108321.
14. Traba J, Miozzo P, Akkaya B, Pierce SK, Akkaya M. An optimized protocol to analyze glycolysis and mitochondrial respiration in lymphocytes. Journal of Visualized Experiments: JoVE. 2016 Nov 21;(117):54918.