Abstract
Due to deregulated control of the cell cycle, proliferation and metabolism, cancer cells are constantly exposed to a wide range of stresses, including DNA damage, nutrient deprivation, heat shock, hypoxia, and oxidative stresses, often, in a non-exclusive manner. Maintaining homeostasis, survival, and proliferation under these adverse conditions is a hallmark of cancer cells. Understanding the factors that enable cancer cells to endure such harsh environments is an important topic in cancer pathology. Recently, the range of these factors have expanded from regulatory proteins, mostly described as oncogenes and tumor suppressors, to non-coding RNAs, particularly long non-coding RNAs (lncRNA). LncRNAs have been found to play crucial roles in regulating stress responses at the molecular level, with implications on cell survival and tumor initiation and progression. In this review, we explore recent studies that underscore the importance of lncRNA in regulating cellular stress responses, homeostasis, and survival.
Keywords
LncRNA, Stress, DNA damage, Hypoxia, Oxidative stress, Metabolic reprograming
Introduction
The human genome is extensively transcribed, producing thousands of non-coding transcripts [1]. Among these, long non-coding RNAs (lncRNA) constitute a major class. LncRNA are typically characterized by their length, exceeding 200 nucleotides. While they resemble mRNAs by being transcribed by RNA polymerase II and sharing similar overall structures, they differ in transcript size, splicing efficiencies, and coding potentials. LncRNA are highly heterogeneous in function and have been shown to regulate various physiological and pathological conditions [2]. Advanced transcriptomics tools have identified numerous dysregulated lncRNA in cancer and other pathological settings [3].
To mitigate the harmful effects of stress, cancer cells utilize all available resources, including both coding and non-coding components of the genome. LncRNAs function as stress-induced-sensors, signal integrators and transducers in physiological and pathological settings. While these activities have been documented, a thorough examination of current literature reveals a lack of recent reviews focusing on the roles of newly discovered lncRNAs in cellular stress management and cell fate determination. This review aims to highlight recent discoveries regarding the involvement of lncRNAs in regulation of UV response, DNA damage signaling, regulation of drug response, oxidative stress, hypoxia and metabolic reprogramming [4-7].
LncRNAs, the UV and DNA Damage Responses (DDR)
Ultraviolet (UV) radiation-driven-damages are one of the most lethal insults that cells may experience in their lifetime [8]. In response to UV exposure, cells execute highly selective and orchestrated gene expression programs to maintain genome integrity and cellular homeostasis, despite a general slowdown in transcript elongation and restriction of gene activity to the promoter-proximal ∼25 kb region [9,10]. While protein-coding genes induced by UV exposure, such as c-Jun, have been well-characterized [11], the involvement of long non-coding RNAs (lncRNAs) in the UV response introduces an additional layer of complexity. For example, the recovery from the global slowdown of transcription in UV-exposed cells is associated with a shift from the expression of long mRNAs to shorter isoforms incorporating alternative last exons (ALEs) that are closer to the transcription start site. One such RNA, ASCC3, shifts from a protein-coding mRNA, which acts as a transcriptional suppressor, to a shorter ALE isoform of non-coding RNA. This shorter isoform, rather than encoding a protein, plays a crucial role in the recovery of transcription [10].
Adaption to environmental stress such as UV-exposure in the organismal level, may also be carried out by the regulation of lncRNAs. For instance, lnc-CD1D-2:1 is upregulated in UVB exposed cells. Its silencing significantly suppressed the UVB induced TYR mRNA expression and tyrosinase activation, suggesting the involvement of lnc-CD1D-2:1 in the melanogenesis induced by UVB irradiation and highlighting the importance of lncRNA for organism’s adaptation to harsh environmental conditions [12].
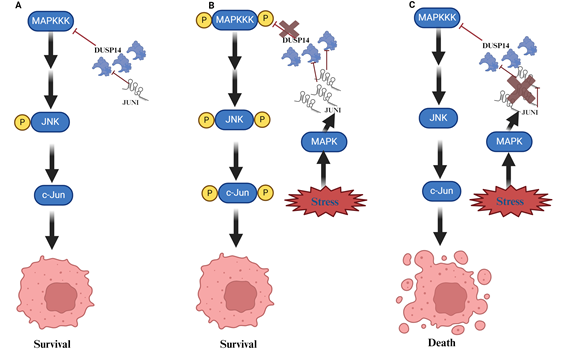
Given the roles of lncRNA in all of the mentioned activities, it is not surprising that they might also influence the fate of UV-exposed cells. We recently demonstrated that the lncRNA JUNI (JUN-DT) is indispensable for the survival of cells exposed to UV. JUNI is located “back-to-back” with the JUN gene and only the JUN promoter distinguishing their transcription start sites. Interestingly, JUNI shares the promoter activity with JUN, a well-established UV responsive gene and like JUN, is positively regulated by MAPK [13]. Expectedly, JUNI is induced by UV exposure and its deficiency significantly sensitizes UV-exposed cells to death as well as decrease JUN expression. In this context, survival is partially dependent on the ability to antagonize MAPK phosphatase DUSP14 (Figure 1).
Figure 1. A cartoon describing the role of JUNI (JUN-DT) for cancer cell survival following UV exposure. A. In normal/unstressed cells, which do not have to cope with stress, the mitogen activated protein kinase (MAPK) pathway, which is composed, among the rest, by c-Jun N-terminal kinase (JNK) and the kinases activating it (MAPKKs and MAPKKKs), is mildly activated, c-Jun is not upregulated. B. In cells exposed to UV the MAPK pathway is activated by phosphorylation cascade, and c-Jun is highly phosphorylated to support survival. MAPK also supports JUNI’s induction that in turn prevents the de-phosphorylation of JNK by interacting and inhibiting the phosphatase DUSP14, thereby enabling efficient induction of c-Jun and survival. C. In cells exposed to UV concomitantly with JUNI (JUN-DT) silencing, DUSP14 activity is not restricted therefore it prevents efficient phosphorylation of JNK and c-Jun activation rendering the cells more sensitive, and increased apoptotic death is observed [21].
The direct involvement of lncRNA in the DNA damage response and DNA repair have been previously reported [14]. Michelini et al., have demonstrated that long non-coding RNAs, termed DilncRNAs, are synthesized by RNA polymerase II (RNAPII) bound to the MRE11-RAD50-NBS1 complex, which is recruited to double strand breaks (DSBs). DilncRNAs act as precursors for DDRNAs and recruit DDRNAs to DSBs through RNA-RNA pairing. Deficiency in DilncRNAs and DDRNAs impairs site-specific DDR focus formation and DNA repair [14]. Another lncRNA, DINO (damage induced), is an important factor for p53 regulated gene expression, cell cycle arrest and cell death post DNA damage [15]. Similarly, the lncRNA HOTTIP is elevated by UV exposure and cooperates with its neighboring gene Hoxa13 to influence UV-induced G2/M-phase arrest and early apoptosis. Mechanistically, both HOTTIP and Hoxa13 regulate the accumulation p53, p21 and γ-H2AX following UV exposure. Interestingly, Hoxa13 overexpression does not significantly affect the cell cycle, apoptosis, or the expression of related proteins in cells not exposed to UV radiation, highlighting the specificity of its activity in UV response [16].
HOTAIR, is a well-studied lncRNA with established involvement in cancer [17]. Upregulation of HOTAIR was found to enhance platinum resistance in ovarian cancer by inducing sustained activation of the DNA damage response (DDR) after platinum treatment and NF-κB activation leading to cellular senescence and drug resistance [18].
LncRNAs-driven Effects on Targeted Therapy and Chemosensitivity
One of the primary reasons for cancer therapy failures is the development of drug resistance [19]. While numerous proteins have been implicated in mediating this phenomenon, recent research has highlighted the significant role of lncRNAs in drug resistance as well. Joung et al., developed a CRISPR-Cas9 based screen that activates over 10,000 distinct lncRNA loci identifying non-coding loci that influence drug resistance. They discovered 11 lncRNA loci that mediate resistance to BRAF inhibitors in human melanoma cells, primarily, by regulating nearby genes [20]. Similarly, the lncRNA JUNI (JUN-DT), which is induced by various chemotherapeutic drugs, has been shown to be a critical regulator of chemosensitivity and cellular survival in cell lines and spheroid models. This effect is likely mediated by the upregulation of its neighboring gene JUN under these conditions [21]. Another lncRNA, HOTAIR, has been found to enhance castration-resistant prostate cancer (CRPC). As its expression is repressed by androgen its levels are upregulated in androgen-depriving- therapies and in CRPC. Mechanistically, HOTAIR binds to the androgen receptor (AR), preventing AR's association with the E3 ubiquitin ligase MDM2. This interaction stabilizes AR, enhances its activity, and thereby increases prostate cancer cell growth and invasion, reducing the efficacy of androgen-deprivation therapies [22]. Linc01126 is also upregulated following androgen deprivation therapy. It regulates AR protein stability and its translocation to the nucleus by modulating O-GlcNAcylation of threonine at position 80 and phosphorylation of serine at position 89. Importantly, overexpression of Linc01126 was sufficient to activate AR signaling in the absence of androgen, while its knockdown led to a significant reduction in resistance, both in vitro and in vivo [23]. In another type of cancer, chronic myeloid leukemia (CML), overexpression of the lncRNA FENDRR mitigates Adriamycin-related resistance, promotes cell death and reduces tumor load in mice [24]. Overall, while research into the involvement of lncRNAs in drug resistance is still in its infancy, the data is promising and warrants further investigation in diverse models to establish their potential benefits in cancer therapy.
LncRNAs and Oxidative Stress
Cancer cells constantly experience oxidative stress, due to higher metabolic activities, oncogenic addiction, genomic instability and drug exposure [25]. Contingent upon concentration, reactive oxygen species (ROS) influence cancer evolution in apparently contradictory ways, either initiating/stimulating tumorigenesis and supporting transformation/proliferation of cancer cells or causing cell death [26]. The expression and activity of lncRNAs have been studied in the context of elevated oxidative stress. Few studies addressed the identification of lncRNA signatures to predict prognosis and immune responses. Zhang et al., identified oxidative-stress-related-lncRNA signature (OSRLs) in gastric cancers, suggesting that eight lncRNAs signature can be used to predict immune response and survival outcome in gastric cancer patients [27]. Similarly, Sun et al., explored the role of lncRNAs associated with oxidative stress in the prognosis and survival of lung adenocarcinoma (LUAD) and identified 16 lncRNAs that can predict the clinical status of LUAD patients [28].
Other studies have found direct involvement of lncRNA in regulating ROS pathways. For example, PVT1, a well-studied lncRNA in cancer has been shown to stabilize NRF2, a key regulator of oxidative stress and promoting resistance to doxorubicin treatment in breast cancer [29]. In contrast, the lncRNA Gas5 increases melanoma cell death and oxidative stress by suppressing translation of EZH2 and increasing the expression of CDKN1C [30].
The lncRNA XIST, known for its essential role in X-chromosome inactivation [31], has been found to be aberrantly overexpressed in non-small cell lung cancer (NSCLC) tumors and cell lines. Knockdown of XIST promotes apoptosis and inhibits cell proliferation in NSCLC cells by inducing pyroptosis. This cell death is associated with increased ROS production and NLRP3 inflammasome activation, and it can be prevented by treatment with the ROS scavenger N-acetyl cysteine (NAC) [32]. Thus, XIST appears to play a non-canonical role in preventing ROS formation and cell death in NSCLC.
Hypoxic and Metabolic Stresses
Hypoxia is a classical feature of tumorigenesis, resulting from uncontrolled cell proliferation and poorly developed, dysregulated vasculature. It has been associated with cancer progression and poor survival outcomes. Previous studies have demonstrated that tumor hypoxia is associated with epithelial-mesenchymal transition, drug resistance, immune evasion, and metabolic reprogramming [33]. Many lncRNAs have been shown to be induced by hypoxia and their roles were delineated [34]. Here, we highlight a few notable examples.
MALAT1, a well-known lncRNA has been studied by several groups in the context of hypoxic conditions in tumors. It was found that MALAT1 inhibits the association of VHL and HIF-1α/HIF-2α, leading to reduced degradation of these hypoxia-inducible factors. This results in increased invasiveness and metastatic potential of arsenite-transformed cells [35,36]. It appears that MALAT1 is not exclusive, the lncRNA CASC9 interacts with HIF-1α, stabilizing it and promoting accelerated glycolysis and tumorigenesis in nasopharyngeal cancer cells [37,38]. HIF-2α itself can directly upregulate the lncRNA NEAT1 in breast cancer cell lines and tumors under hypoxic conditions. This upregulation leads to increased paraspeckle formation and consequently to sequestration of proteins and RNAs, revealing a novel mechanism of gene regulation by lncRNAs under hypoxia. Elevated NEAT1 expression is associated with higher proliferation and survival in cancer cell lines, as well as poor prognosis in patients [39].
The cytoplasmic lncRNA LINK-A has been shown to activate HIF-1α transcriptional programs under normoxic conditions. LINK-A recruits BRK to the EGFR complex, leading to BRK phosphorylation and stabilization of HIF-1α. This LINK-A-dependent signaling is correlated with triple-negative breast cancer (TNBC) and promotes glycolysis reprogramming in breast cancer [40]. Additionally, the TNBC cells MDA-MB-231 acquire stem cell-like features through the regulation of the SETDB1/PLK3/HIF1α signaling pathway by linc00115. This lncRNA is significantly upregulated following paclitaxel treatment, contributing to drug resistance [41].
Metabolic stress is a well-documented phenomenon in cancer [42] and in fact, metabolic changes are a driving force in some cancers, such as kidney cancer [43]. The prevailing involvement of lncRNA in response to stresses is also correct for the accommodation of metabolic ones. The mitochondrial lncRNA GAS5 is a critical regulator of TCA cycle and metabolic stress, which interacts with malate dehydrogenase 2 (MDH2), an important factor for citrate synthase complex formation and cancer progression [44].
Glucose consumption, an extremely important factor for cancer cell survival is also regulated by lncRNAs. MALAT1, for instance, contributes to hepatocellular carcinoma (HCC) development and tumor progression by reprogramming tumor glucose metabolism. Mechanistically, it enhances the translation of the metabolic transcription factor TCF7L2, in mTOR dependent manner [45].
LncRNAs also regulate the execution of hypoxic programs under normoxic conditions, enhancing tumorigenicity. For example, the lncRNA, IDH1-AS1, prevents the formation of Warburg effect under normoxia whereas c-Myc transcriptionally represses IDH1-AS1 to enable Warburg effect under normoxia [46]. Additionally, the lncRNA SOX9-AS1 affects lipid metabolism in breast cancer cells. Cisneros-Villanueva et al., discovered that the lncRNA SOX9-AS1 is significantly overexpressed in basal-like and TNBC breast tumors compared to other breast cancer tumor types. Its overexpression was associated with favorable prognosis in TNBC and basal-like patients. Furthermore, the researchers found that lncRNA SOX9-AS1 promoted lipid metabolism in these cancer types reducing triglyceride synthesis, cell migration and invasion in two TNBC cell lines. Its suppression enhances aggressiveness of TNBC cells [47]. Moreover, lncRNAs can also regulate the function of NAD+ dependent SIRT1 histone deacetylase, a highly conserved and well-studied cytoprotective protein against various cellular insults such as oxidative stress, DNA damage, and metabolic reprogramming [48,49]. For instance, Zhang et al., for example, demonstrated that the lncRNA GAS5, an lncRNA also involved in various metabolic effects, inhibits malignant properties in colon cancer by regulating SIRT1 [50].
Conclusion and Future Directions
Cancer thrives under stressful conditions and constantly adapts by exploiting both coding and non-coding genes. Among non-coding RNAs, long non-coding RNAs (lncRNAs) play a crucial role in responding to cellular stress and maintaining homeostasis. Numerous studies have highlighted the involvement of lncRNAs in regulating stress responses related to specific organelles, cytoplasmic-nuclear communication, genotoxic stress, and hyperactivated molecular pathways during cancer progression and therapy [51,52].
However, many lncRNA were overlooked due to lower expression and higher variability across different tissues. To address this gap, a deeper understanding of differential lncRNAs expression and function in different disease models is required. Mapping the full spectrum of lncRNAs that enable cancer cells to cope with the endogenous stresses or those induced by specific therapies is necessary. This includes studying lncRNAs with lower expression levels or those critical to specific stages of the cell cycle, such as cell division distinguishing cancer cells from the background of non-proliferating normal tissue. Thereby, expending the “Target bank” in the fight against different types of cancers.
The identification, using various biochemical and bioinformatics tools as well as hypothesis-driven experiments, of genes, motifs, secondary and tertiary structures for better understanding of lncRNA activity under stressful conditions are warranted, but remains in a primary stage. Therapies exploiting the essentiality of such factors, targeting them as monotherapy or in combination with stress inducing agents, will be possible only upon development of better nucleic-acid-based delivery systems for therapy.
References
2. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 2023 Jun;24(6):430-47.
3. Delás MJ, Hannon GJ. lncRNAs in development and disease: from functions to mechanisms. Open Biol. 2017 Jul;7(7):170121.
4. Liu X, Feng S, Zhang XD, Li J, Zhang K, Wu M, et al. Non-coding RNAs, metabolic stress and adaptive mechanisms in cancer. Cancer Lett. 2020 Oct 28;491:60-9.
5. Connerty P, Lock RB, de Bock CE. Long Non-coding RNAs: Major Regulators of Cell Stress in Cancer. Front Oncol. 2020 Mar 20;10:285.
6. Park EG, Pyo SJ, Cui Y, Yoon SH, Nam JW. Tumor immune microenvironment lncRNAs. Brief Bioinform. 2022 Jan 17;23(1):bbab504.
7. Nickerson JA, Momen-Heravi F. Long non-coding RNAs: roles in cellular stress responses and epigenetic mechanisms regulating chromatin. Nucleus. 2024 Dec;15(1):2350180.
8. Anzi S, Finkin S, Shaulian E. Transcriptional repression of c-Jun's E3 ubiquitin ligases contributes to c-Jun induction by UV. Cell Signal. 2008 May;20(5):862-71.
9. Shaulian E, Schreiber M, Piu F, Beeche M, Wagner EF, Karin M. The mammalian UV response: c-Jun induction is required for exit from p53-imposed growth arrest. Cell. 2000 Dec 8;103(6):897-907.
10. Williamson L, Saponaro M, Boeing S, East P, Mitter R, Kantidakis T, et al. UV Irradiation Induces a Non-coding RNA that Functionally Opposes the Protein Encoded by the Same Gene. Cell. 2017 Feb 23;168(5):843-55.e13.
11. Angel P, Allegretto EA, Okino ST, Hattori K, Boyle WJ, Hunter T, et al. Oncogene jun encodes a sequence-specific trans-activator similar to AP-1. Nature. 1988 Mar 10;332(6160):166-71.
12. Zeng Q, Wang Q, Chen X, Xia K, Tang J, Zhou X, et al. Analysis of lncRNAs expression in UVB-induced stress responses of melanocytes. J Dermatol Sci. 2016 Jan;81(1):53-60.
13. Kumar V, Sabaté-Cadenas X, Soni I, Stern E, Vias C, Ginsberg D, et al. The lincRNA JUNI regulates the stress-dependent induction of c-Jun, cellular migration and survival through the modulation of the DUSP14-JNK axis. Oncogene. 2024 May;43(21):1608-19.
14. Michelini F, Pitchiaya S, Vitelli V, Sharma S, Gioia U, Pessina F, et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat Cell Biol. 2017 Dec;19(12):1400-11.
15. Schmitt AM, Garcia JT, Hung T, Flynn RA, Shen Y, Qu K, et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet. 2016 Nov;48(11):1370-6.
16. Liang M, Hu K. Involvement of lncRNA-HOTTIP in the Repair of Ultraviolet Light-Induced DNA Damage in Spermatogenic Cells. Mol Cells. 2019 Nov 30;42(11):794-803.
17. Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015 Mar;12(1):1-9.
18. Özeş AR, Miller DF, Özeş ON, Fang F, Liu Y, Matei D, et al. NF-κB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016 Oct 13;35(41):5350-61.
19. Lau MT, Ghazanfar S, Parkin A, Chou A, Rouaen JR, Littleboy JB, et al. Systematic functional identification of cancer multi-drug resistance genes. Genome Biol. 2020 Feb 7;21(1):27.
20. Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017 Aug 17;548(7667):343-6
21. Kumar V, Sabaté-Cadenas X, Soni I, Stern E, Vias C, Ginsberg D, et al. The stress-induced lincRNA JUNI is a critical factor for cancer cell survival whose interactome is a prognostic signature in clear cell renal cell carcinoma. bioRxiv. 2023:2023-10.
22. Zhang A, Zhao JC, Kim J, Fong KW, Yang YA, Chakravarti D, et al. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep. 2015 Oct 6;13(1):209-21.
23. Cai Y, Chen M, Gong Y, Tang G, Shu Z, Chen J, et al. Androgen-repressed lncRNA LINC01126 drives castration-resistant prostate cancer by regulating the switch between O-GlcNAcylation and phosphorylation of androgen receptor. Clin Transl Med. 2024 Jan;14(1):e1531.
24. Zhang F, Ni H, Li X, Liu H, Xi T, Zheng L. LncRNA FENDRR attenuates adriamycin resistance via suppressing MDR1 expression through sponging HuR and miR-184 in chronic myelogenous leukaemia cells. FEBS Lett. 2019 Aug;593(15):1993-2007.
25. Scandalios JG. Oxidative stress responses--what have genome-scale studies taught us? Genome Biol. 2002 Jun 18;3(7):REVIEWS1019.
26. Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer cell. 2020 Aug 10;38(2):167-97.
27. Zhang H, Feng H, Yu T, Zhang M, Liu Z, Ma L, et al. Construction of an oxidative stress-related lncRNAs signature to predict prognosis and the immune response in gastric cancer. Sci Rep. 2023 May 31;13(1):8822.
28. Sun X, Huang X, Sun X, Chen S, Zhang Z, Yu Y, et al. Oxidative Stress-Related lncRNAs Are Potential Biomarkers for Predicting Prognosis and Immune Responses in Patients With LUAD. Front Genet. 2022 Jun 8;13:909797.
29. Luo Y, Zhang W, Xu L, Chen Y, Xu Y, Yuan L. Long Non-Coding RNA PVT1 Regulates the Resistance of the Breast Cancer Cell Line MDA-MB-231 to Doxorubicin via Nrf2. Technol Cancer Res Treat. 2020 Jan-Dec;19:1533033820980763.
30. Xu W, Yan Z, Hu F, Wei W, Yang C, Sun Z. Long non-coding RNA GAS5 accelerates oxidative stress in melanoma cells by rescuing EZH2-mediated CDKN1C downregulation. Cancer Cell Int. 2020 Apr 9;20:116.
31. Marahrens Y, Loring J, Jaenisch R. Role of the Xist gene in X chromosome choosing. Cell. 1998 Mar 6;92(5):657-64.
32. Liu J, Yao L, Zhang M, Jiang J, Yang M, Wang Y. Downregulation of LncRNA-XIST inhibited development of non-small cell lung cancer by activating miR-335/SOD2/ROS signal pathway mediated pyroptotic cell death. Aging (Albany NY). 2019 Sep 25;11(18):7830-46.
33. Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl). 2015 Dec 11;3:83-92.
34. Peng PH, Hsu KW, Chieh-Yu Lai J, Wu KJ. The role of hypoxia-induced long noncoding RNAs (lncRNAs) in tumorigenesis and metastasis. Biomed J. 2021 Oct;44(5):521-33.
35. Luo F, Sun B, Li H, Xu Y, Liu Y, Liu X, et al. A MALAT1/HIF-2α feedback loop contributes to arsenite carcinogenesis. Oncotarget. 2016 Feb 2;7(5):5769-87.
36. Luo F, Liu X, Ling M, Lu L, Shi L, Lu X, et al. The lncRNA MALAT1, acting through HIF-1α stabilization, enhances arsenite-induced glycolysis in human hepatic L-02 cells. Biochim Biophys Acta. 2016 Sep;1862(9):1685-95.
37. Su X, Li G, Liu W. The Long Noncoding RNA Cancer Susceptibility Candidate 9 Promotes Nasopharyngeal Carcinogenesis via Stabilizing HIF1α. DNA Cell Biol. 2017 May;36(5):394-400.
38. Zhang Z, Fang E, Rong Y, Han H, Gong Q, Xiao Y, et al. Hypoxia-induced lncRNA CASC9 enhances glycolysis and the epithelial-mesenchymal transition of pancreatic cancer by a positive feedback loop with AKT/HIF-1α signaling. Am J Cancer Res. 2021 Jan 1;11(1):123-37.
39. Choudhry H, Albukhari A, Morotti M, Haider S, Moralli D, Smythies J, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015 Aug 20;34(34):4482-90.
40. Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. 2016 Feb;18(2):213-24.
41. Luo F, Zhang M, Sun B, Xu C, Yang Y, Zhang Y, et al. LINC00115 promotes chemoresistant breast cancer stem-like cell stemness and metastasis through SETDB1/PLK3/HIF1α signaling. Mol Cancer. 2024 Mar 22;23(1):60.
42. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012 Mar 20;21(3):297-308.
43. Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol. 2010 May;7(5):277-85.
44. Sang L, Ju HQ, Yang Z, Ge Q, Zhang Z, Liu F, et al. Mitochondrial long non-coding RNA GAS5 tunes TCA metabolism in response to nutrient stress. Nat Metab. 2021 Jan;3(1):90-106.
45. Malakar P, Stein I, Saragovi A, Winkler R, Stern-Ginossar N, Berger M, et al. Long Noncoding RNA MALAT1 Regulates Cancer Glucose Metabolism by Enhancing mTOR-Mediated Translation of TCF7L2. Cancer Res. 2019 May 15;79(10):2480-93.
46. Xiang S, Gu H, Jin L, Thorne RF, Zhang XD, Wu M. LncRNA IDH1-AS1 links the functions of c-Myc and HIF1α via IDH1 to regulate the Warburg effect. Proc Natl Acad Sci U S A. 2018 Feb 13;115(7):E1465-E1474.
47. Cisneros-Villanueva M, Fonseca-Montaño MA, Ríos-Romero M, López-Camarillo C, Jiménez-Morales S, Langley E, et al. LncRNA SOX9-AS1 triggers a transcriptional program involved in lipid metabolic reprogramming, cell migration and invasion in triple-negative breast cancer. Sci Rep. 2024 Jan 17;14(1):1483.
48. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021 Feb;22(2):96-118.
49. Ghafouri-Fard S, Shoorei H, Hussen BM, Poornajaf Y, Taheri M, Sharifi G. Interaction between SIRT1 and non-coding RNAs in different disorders. Front Genet. 2023 Jun 27;14:1121982.
50. Zhang HG, Wang FJ, Wang Y, Zhao ZX, Qiao PF. lncRNA GAS5 inhibits malignant progression by regulating macroautophagy and forms a negative feedback regulatory loop with the miR 34a/mTOR/SIRT1 pathway in colorectal cancer. Oncol Rep. 2021 Jan;45(1):202-16.
51. Sekine Y, Houston R, Sekine S. Cellular metabolic stress responses via organelles. Exp Cell Res. 2021 Mar 1;400(1):112515.
52. Larini L, Goretti E, Zulian E, Busarello E, Marino SM, Hajikazemi M, et al. Telomeric lncRNA TERRA localizes to stress granules in human ALT cells. bioRxiv. 2024:2024-06.