Abstract
OAS1 plays a critical role in host-pathogen interactions by balancing translational shutdown to limit microbial replication while producing antimicrobial components. Host RNA sensors detect microbial nucleic acids, initiating an interferon-mediated innate immune response that induces ISGs to inhibit replication and shape adaptive immunity. OAS1 enhances the translation of selective mRNAs, producing proteins with antimicrobial properties. During infections, OAS1, induced by interferons, identifies viral RNA, and responds to viruses that replicate within modified organelles. Genetic polymorphisms influence OAS1’s function, affecting susceptibility to various infections. The recent study by Harioudh et al. reveals OAS1's mechanism in enhancing mRNA translation to boost antiviral and antibacterial activities, involving pathways like cGAS-STING and IRF1. OAS1's roles in autoimmune diseases and cancer, and its dual role in immune activation and potential cellular damage, highlight the need for further research to harness its therapeutic potential while mitigating adverse effects.
Keywords
Oligoadenylate Synthase 1 (OAS1), Interferon, Antiviral, Antibacterial, Immunity, Immunomodulation
Graphical Abstract
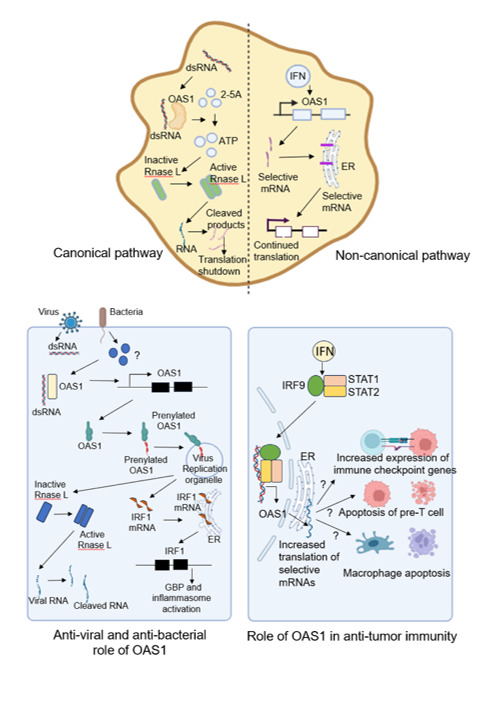
The graphical abstract illustrates the canonical and non-canonical pathway of OAS1 activation. Also, the antiviral, antibacterial, and anti-tumorigenic role of OAS1 activity is schematically represented in the figure.
Commentary
In the context of host-pathogen interaction, the host faces the challenge of balancing translational shutdown to limit microbial replication while maintaining the production of antimicrobial components. Host RNA sensors detect nucleic acids from invading microbes, initiating innate immune response mediated by interferons. These interferons induce the expression of interferon-stimulated genes (ISGs) to inhibit microbial replication and shape the adaptive immune system to clear the infection. Endomembrane targeting of the innate immune sensors and translational reprogramming via location-specific translation of selective mRNAs represent the survival strategies for the host.
Previous studies have highlighted the role of an ISG, Oligoadenylate synthase 1 (OAS1) in combating viral and bacterial infections. However, the precise molecular mechanisms underlying this interferon-dependent antimicrobial response remained unclear. In a recent Immunity issue, Harioudh et al., reported a unique mechanism by which OAS1 binds and enhances the translation of selective mRNAs, thereby inducing the production of specific proteins with antimicrobial properties [1]. This commentary summarizes the current understanding of the role of OAS1 in antiviral and antibacterial responses and explores how these attributes could be leveraged for immunomodulation in cancer and auto-immune disorders.
During viral infections, OAS1 is induced by interferons to locate the RNA viruses in their concealed locations. Host RNA sensors, typically positioned in the cytosol detect viral RNA during translation. OAS1, constitutively expressed, acts as a pattern recognition receptor (PRR) to identify viral RNAs in the cytoplasm [2]. Viruses have evolved to replicate within modified organelles derived from the endomembrane system, shielding themselves from innate immune responses. In response, the host has co-evolved shaping their OAS1 repertoires with polymorphisms that position it on the endomembrane system [3]. The question “What targets OAS1 to the replication organelles of RNA viruses?” was addressed in a remarkable study by Wickenhagen et al., published in Science, 2021. They reported prenylations of OAS1 are associated with the severity of COVID-19. Prenylated OAS1 can infiltrate viral replication organelles, detect ‘hidden’ viruses, and mount an antiviral immune response [4].
To counteract virus infections, interferons induce the expression of ISGs, including 2’, 5’-oligoadenylate synthetase 1(OAS1). As a part of the innate immune response, OAS1 plays a key role in recognizing viral dsRNA and activating RNase L, leading to the degradation of viral RNA via an RNA decay pathway [5]. This process appears quite straightforward but cells must distinguish between “self” and “non-self” RNAs to avoid constitutive antiviral responses.
A noteworthy discovery by Calderon et al., demonstrated that ‘shape-shifting’ RNAs, including human noncoding RNA 886(nc886), contribute to this discrimination [6], although the detailed mechanism remains unclear. Recent studies have also identified several RNase L-independent pathways through which OAS proteins exert antiviral activities [1]. Single nucleotide polymorphisms in the OAS1 gene can affect its expression, enzymatic activity, and alternative splicing, influencing susceptibility to various viral infections [7]. For instance, SNP rs2660 (A→G) near the exon 7 splice-acceptor site of the OAS1 gene alters 2-5OAS1 enzyme activity [8]. The AA genotype is associated with low enzymatic activity, while the GG genotype confers high enzymatic activity [9]. These genetic variations are linked to susceptibility to hepatitis C [10], influenza A [11], flavivirus [12], West Nile virus [13], respiratory epithelial cell syncytial virus [14], ocular herpes simplex virus type 1, and severe acute respiratory syndrome (SARS) [15].
OAS1 is linked to susceptibility to M. tuberculosis and other infections through various single nucleotide polymorphisms (SNPs), such as rs10774671 [16]. This SNP, which influences susceptibility to Hepatitis B and West Nile virus also affects bacterial infection susceptibility Harioudh et al. revealed a novel mechanism where OAS1 enhances the translation of selective mRNAs thereby boosting antiviral and antibacterial activities [1]. Their study showed that OAS1 enhanced cGAS expression, restricting WNV via the cGAS-STING pathway. Using L. monocytogenes (Lm) and F. novicida (Fn) as bacterial models, they discovered that the same SNP associated with WNV resistance also restricts M. tuberculosis, suggesting a shared protective mechanism. The antibacterial role of OAS1 involves the downstream protein IRF1. OAS1-deficient cells exhibited increased bacterial growth when infected with Lm and Fn, while inducible expression of P46 isoform of OAS1 significantly reduced bacterial burden. This antibacterial activity was abolished in IRF1-deficient cells, highlighting IRF1’s essential role. Transcriptomic and proteomic analysis revealed that OAS1 regulates several transcription factors post-transcriptionally, including GBPs. The study demonstrated that OAS1 localizes to the ER directly binds to mRNAs, and enhances their translation, promoting the synthesis of basal and IFN-inducible proteins such as cGAS and IRF1. This leads to type I IFN signalling necessary for infection clearance and the activation of innate immune genes like GBPs, which disrupt bacteria-containing vacuoles or affect bacterial membrane integrity. In vivo experiments with WT and Oas1b-KI mice infected with Lm and Fn showed that Oas1b-KI mice had, significantly higher survival rates. Current projects in the author's lab are to further elucidate the role of GBPs and inflammasome signalling on antibacterial immunity.
‘A good offense doesn’t always prove to be a good strategy in host-pathogen interactions!’ Despite the clear benefits demonstrated by innate immunity to host fitness, it can also incur costs in the absence of the ‘invading enemy’. In an issue of Cell Host Microbe, Clayton et al. revealed that OAS1 autoactivation by host RNAs can serve as a trade-off to its antiviral activity, by disrupting cellular RNAs, while sparing mRNAs encoding interferons and ISGs. Several groups hypothesized that selective expression of innate immune mRNAs and proteins combined with the translational arrest of housekeeping and metabolic genes may create a positive feedback loop, thereby stoking an inflammatory immune response [17]. In an issue of Science Immunology, Thomas et al., reported OAS1 gain-of-function variants to be associated with autoinflammatory immunodeficiency thereby highlighting the cost-benefit conundrum of immune functions on host fitness, where deleterious effects of immune activation may sometimes outweigh the benefits of host defense [18].
Upregulated OAS1 expression is found in breast and pancreatic cancer patients [19], thereby serving as a prognostic biomarker for several tumor types. These findings raised a huge questionnaire on the correlation of OAS1 expression with the expression of immune checkpoint genes and tumor burden. OAS1 SNP rs2660 AA genotype is associated with prostate cancer. Gain-of-function variants of OAS1 are associated with apoptosis of macrophages in autoimmune diseases [18] but the exact correlation of OAS1 and M2 macrophages are still under investigation. Studies report an association of OAS1 with T-cell dysfunction and macrophage M2 infiltration [20], but the underlying mechanisms are still unclear. A significant knowledge gap exists in understanding the role of OAS1 in tumor immune evasion and immunotherapeutic resistance. OAS1 mediates apoptosis of thymic pre-T cells and B-cells having increased OAS1 expression are prone to apoptosis. Monocytes and B cells were found to be susceptible to OAS1 activation [18,21]. These findings raise the question, ‘Is OAS1 involved in shaping the B and T cell repertoires?’ In line with this evidence, several groups speculate that OAS1 might constitute a cell-intrinsic immune checkpoint. Can OAS1 activation and subsequent downstream signaling balance the need for antigen presentation and antigen spreading by ‘deleting cells’ that exceed a signaling threshold induced by foreign dsRNA? Another noteworthy observation was reported by Daniel et al. in Science Immunology, where the authors reported OAS1 to be involved in tumor cell apoptosis via translational arrest and depletion of anti-apoptotic MCL-1 [22]. A parallel study published in EmboJ by Anna et al., revealed OAS1 as an interferon-related DNA damage resistance signature, thereby indicating its role as ‘safety switches’ in tumor immunity [23].
Questions and Future Perspectives
1. Viral nucleic acids are sensed by OAS proteins so what is the nature of bacterial product ‘seen’ by the innate immune RNA sensors?
2. Can cells combat bacteria when prompted to counter virus infections?
3. Owing to the various polymorphisms in the OAS gene family, which accounts for their various anti-microbial functions, how does the host shape their OAS repertoires to trick the virus-evolved avoidance of antiviral effectors?
4. How is the IRF1 mRNA transported to the endomembrane upon binding OAS1?
5. As OAS1 is upregulated in virus and bacterial infections, can OAS1 levels be used as biomarkers for detecting infections and monitoring the effect of therapy?
6. Owing to the perceived effect of OAS1 on RNase L and the fact that RNase L deficient cells are impaired in their ability to induce apoptosis, can OAS1 have non-immunological functions like regulation of apoptosis to eliminate virus-infected cells?
7. As OAS1 has been reported to be a prognostic biomarker in several cancers via clinical studies and IRF1 is a Tumor suppressor gene, can OAS1-mediated increased translation of IRF1 mRNA provide anti-Tumor immunity?
8. Owing to the fact that OAS1 causes disruption of cellular RNA, how do the cells protect themselves from the deleterious effects of OAS1 activation in the absence of infection? Does the editing of cellular RNA suppress the RNase L-induced lethality?
9. Does OAS1 play any role in shaping the adaptive immune repertoire? Further insights into the role of OAS proteins in adaptive immunity would open avenues to manipulate immunity to enhance resistance to pathogens.
Conclusion
In conclusion, the intricate interplay between OAS1 and host-pathogen interactions highlights its pivotal role in innate immunity. OAS1, activated by interferons during infections, orchestrates a multifaceted defense strategy by enhancing the translation of select mRNAs, crucial for combating both viral and bacterial pathogens. Recent studies have elucidated its involvement in RNA sensing, regulation of antiviral and antibacterial responses through pathways like cGAS-STING and IRF1, and implications in autoimmune diseases and cancer. While offering promising therapeutic avenues, including immunomodulation in cancer, OAS1's dual role in immune activation and potential cellular damage underscores the need for further research to harness its benefits effectively while minimizing unintended consequences on host physiology.
Funding
No financial support is involved.
Acknowledgment
SG, SD acknowledge the support from CSIR-IICB; SD acknowledges the Department of Biotechnology for supporting her as a DBT-JRF Fellow, ID - DBT/2022-23/IICB/1982 and Academy of Scientific and Innovative Research (AcSIR), Ghaziabad- 201002, India.
Author Contributions
SG conceived the theme of the commentary; SG and SD performed the literature search and composed the commentary and graphics. The authors also acknowledge Biorender.com as the platform used for the illustration of the figure.
Conflicts of Interest
The authors declare no conflict of interest and the sponsors had no role in the design, execution, interpretation, or writing of the study.
References
2. Schwartz SL, Park EN, Vachon VK, Danzy S, Lowen AC, Conn GL. Human OAS1 activation is highly dependent on both RNA sequence and context of activating RNA motifs. Nucleic Acids Res. 2020 Jul 27;48(13):7520-31.
3. Soveg FW, Schwerk J, Gokhale NS, Cerosaletti K, Smith JR, Pairo-Castineira E, et al. Endomembrane targeting of human OAS1 p46 augments antiviral activity. Elife. 2021 Aug 3;10:e71047.
4. Wickenhagen A, Sugrue E, Lytras S, Kuchi S, Noerenberg M, Turnbull ML, et al. A prenylated dsRNA sensor protects against severe COVID-19. Science. 2021 Oct 29;374(6567):eabj3624.
5. Harioudh MK, Perez J, Chong Z, Nair S, So L, McCormick KD, et al. Oligoadenylate synthetase 1 displays dual antiviral mechanisms in driving translational shutdown and protecting interferon production. Immunity. 2024 Mar 12;57(3):446-61.e7.
6. Calderon BM, Conn GL. A human cellular noncoding RNA activates the antiviral protein 2'-5'-oligoadenylate synthetase 1. J Biol Chem. 2018 Oct 12;293(41):16115-24.
7. Banday AR, Stanifer ML, Florez-Vargas O, Onabajo OO, Zahoor MA, Papenberg BW, et al. Genetic regulation of OAS1 nonsense-mediated decay underlies association with risk of severe COVID-19. medRxiv [Preprint]. 2021 Jul 13:2021.07.09.21260221.
8. Mandal SK, Abebe F, Chaudhary J. 2’-5’oligoadenylate synthetase polymorphism is associated with prostate cancer: Effect modified by age and race. Cancer Research. 2011 Apr 15;71(8_Supplement):2772.
9. O'Brien M, Lonergan R, Costelloe L, O'Rourke K, Fletcher JM, Kinsella K, et al. OAS1: a multiple sclerosis susceptibility gene that influences disease severity. Neurology. 2010 Aug 3;75(5):411-8.
10. Knapp S, Yee LJ, Frodsham AJ, Hennig BJ, Hellier S, Zhang L, et al. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun. 2003 Sep;4(6):411-9.
11. Min JY, Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2'-5' oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci U S A. 2006 May 2;103(18):7100-5.
12. Urosevic N. Is flavivirus resistance interferon type I-independent? Immunol Cell Biol. 2003 Jun;81(3):224-9.
13. Yakub I, Lillibridge KM, Moran A, Gonzalez OY, Belmont J, Gibbs RA, et al. Single nucleotide polymorphisms in genes for 2'-5'-oligoadenylate synthetase and RNase L inpatients hospitalized with West Nile virus infection. J Infect Dis. 2005 Nov 15;192(10):1741-8.
14. Behera AK, Kumar M, Lockey RF, Mohapatra SS. 2'-5' Oligoadenylate synthetase plays a critical role in interferon-gamma inhibition of respiratory syncytial virus infection of human epithelial cells. J Biol Chem. 2002 Jul 12;277(28):25601-8.
15. Hamano E, Hijikata M, Itoyama S, Quy T, Phi NC, Long HT, et al. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem Biophys Res Commun. 2005 Apr 22;329(4):1234-9.
16. Wu S, Wang Y, Chen G, Zhang M, Wang M, He JQ. 2'-5'-Oligoadenylate synthetase 1 polymorphisms are associated with tuberculosis: a case-control study. BMC Pulm Med. 2018 Nov 29;18(1):180.
17. Carey CM, Govande AA, Cooper JM, Hartley MK, Kranzusch PJ, Elde NC. Recurrent Loss-of-Function Mutations Reveal Costs to OAS1 Antiviral Activity in Primates. Cell Host Microbe. 2019 Feb 13;25(2):336-43.e4.
18. Magg T, Okano T, Koenig LM, Boehmer DFR, Schwartz SL, Inoue K, et al. Heterozygous OAS1 gain-of-function variants cause an autoinflammatory immunodeficiency. Sci Immunol. 2021 Jun 18;6(60):eabf9564.
19. Jiang S, Deng X, Luo M, Zhou L, Chai J, Tian C, et al. Pan-cancer analysis identified OAS1 as a potential prognostic biomarker for multiple tumor types. Front Oncol. 2023 Sep 6;13:1207081.
20. Yang R, Du Y, Zhang M, Liu Y, Feng H, Liu R, et al. Multi-omics analysis reveals interferon-stimulated gene OAS1 as a prognostic and immunological biomarker in pan-cancer. Front Immunol. 2023 Oct 20;14:1249731.
21. Lu L, Wang H, Fang J, Zheng J, Liu B, Xia L, et al. Overexpression of OAS1 Is Correlated With Poor Prognosis in Pancreatic Cancer. Front Oncol. 2022 Jul 11;12:944194.
22. D Böhmer S, Formisano C, Mann S, Müller M, Kluge P, Metzger M, et al. OAS1/RNase L executes RIG-I ligand–dependent tumor cell apoptosis, Sci Immunol 2021 Jul 16;6(61):eabe2550.
23. Kondratova AA, Cheon H, Dong B, Holvey-Bates EG, Hasipek M, Taran I, et al. Suppressing PARylation by 2',5'-oligoadenylate synthetase 1 inhibits DNA damage-induced cell death. EMBO J. 2020 Jun 2;39(11):e101573.
