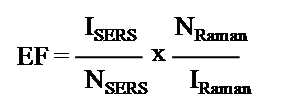Abstract
Preparation of nanomaterials (NMs) using bio-molecules such as DNA as scaffolds is a simple, greener, and reliable way of designing the size and shape selective NMs. The Rh nanoparticles prepared on DNA scaffolds generate perfect nano-networks structure by utilizing a simple wet-chemical method in presence of NaBH4 as reducing agent. The prepared Rh nanoparticles on DNA scaffolds were effectively utilized for Surface-Enhanced Raman Scattering (SERS) studies using methylene blue (MB) as a SERS probe molecule. The DNA facilities to control the Rh NPs to grow below 5 nm in diameter and effectively utilized to increase the Raman signal. The highest SERS enhancement factor (EF) calculated was 1.19×105 for the lowest probe detection limit of 10-6 M. The stability of these Rh nano-networks were verified by aging the sample for more than six months which shows the same SERS efficiency.
Keywords
Rh Nano-networks; Biomolecule; DNA; Hotspot; Enhancement factor Rh Nano-networks; Biomolecule; DNA; Hotspot; Enhancement factor Rh Nano-networks; Biomolecule; DNA; Hotspot; Enhancement factor
Introduction
The study of surface and interfacial science via analytical techniques is a growing interest in the easy mode of diagnosing probe molecules adsorbed on the surface [1]. In that line, Raman spectroscopy is an analytical method that is inefficient to carry the surface analysis employing their weak cross-sectional scattering until the discovery of Surface-Enhanced Raman Scattering (SERS) in late 1970s [2,3]. SERS creates a revolution in many fields, including sensor and biomedical applications [4-6]. It works, when the incident light hits on rough metal surface adsorbed on the probe molecule gets increased Raman signal in a few orders of magnitude [7,8]. The noble metal-based nanomaterials (NMs) such as Ag [9], Au [10], Pt [11], Pd [12], Re [13], Os [14], Ir [15], and RuO2[16] having the remarkable surface plasmon resonance (SPR) nature in the visible region. Many factors that are influencing the enhancement of such SERS signal mainly, wavelength, polarization, and incidence angle of the laser light [17-19]. Besides, the size and shape of nanomaterials was greatly provoking the SERS signal by increasing the hot-spots [ 20]. Therefore, tuning of such NMs in a controlled way of keeping the inter-particle gap below 5 nm (more hotspots) is a remarkable way of increasing the SERS signal [9]. To increase such hot spots in the NMs using simple methods is the ultimate aim of the researchers to get a high enhancement factor [21-23]. Noble metals such as Ag, Au, and Pd has been well studied earlier [9,24], which are more active contenders for SERS. In that line, Rh NMs is a less explored material as SERS substrate for probe detection [25]. Thus, we have chosen Rh NPs prepared on bio-molecule DNA as scaffold for an active SERS substrate to carry the probe detection.
Owing to its wide tune ability and green nature, biomolecules based NMs synthesis is a growing interest among the scientific community [26]. In such a way, deoxyribonucleic acid (DNA) is a best choice of preparing NMs and its diameter is 2 nm and the length is 0.34 nm [27]. Thus, it perfectly meets the same line with the material science [28]. Also, the double-helix structure contains nucleotide base pairs of adenine (A), guanine (G), thymine (T) and cytosine (C) were connected by the hydrogen bonding [29]. The presence of functionalities in DNA such as phosphate groups and sugar moieties are more facile to accommodate metal ions via electrostatic interaction [30]. Therefore, fabricating closely packed nanomaterial to ensure more hot-spots is highly desirable using the biomolecule DNA [9]. In addition, the presence of DNA can stabilize the NMs in the aqueous solution over long period [15,31]. At first, the metal ion makes an electrostatic interaction with the negative moieties in the double-helix of DNA [26,32-34]. Next to this activation stage, the reducing agents used to convert reduce these metal ions to metallic state [35]. At this stage, the NMs starts to nucleate on the DNA over and over the surface like a nucleation site and forms a perfect nano-networks [ 35-38].
In this commentary, we are reporting a facile way of synthesising Rh nano-networks using a bio-molecular scaffolds such as DNA. The overall reaction took 40 min to complete and the synthesized Rh NPs on DNA are stable for long time (more than six months). Also, the observed nano-networks diameter was calculated to be ~ 20 nm and the individual particle size was below 5 nm. These Rh nano-networks were highly active for SERS studies for the probe detection of Methylene Blue (MB). The synthesis process is simple, reproducible, less-time consuming and occurs at room temperature.
Experimental Section
Reagents and the instruments used
The materials such as Rhodium chloride (RhCl3.H2O) (98%) and Sodium Borohydride (NaBH4) (99%) were obtained from Sigma-Aldrich and used as received. Double stranded Deoxyribonucleic acid (DNA) (99%) from Herring Testes with a base pair of around 50 k has been received from Sigma-Aldrich and used directly for the preparation of stock solution. The dye molecule methylene blue (MB) was purchased from Qualigens Fine Chemicals (99%), Mumbai. Milli-Q water was used for the complete reactions and sample preparations. The X-ray diffraction (XRD) analysis was carried by using a PAN analytical Advanced Bragg-Brentano X-ray powder diffractometer (XRD) with Cu Kα radiation (λ = 0.154178 nm) with a scanning rate of 0.020 s-1 in the 2θ range 10-90°. The UVVisible absorption (UV-Vis) spectra were carried in a Unico (model 4802) UV-Vis-NIR spectrophotometer equipped with a 1 cm quartz cuvette holder mainly for liquid samples. The morphological studies and the HAADF color mapping of Rh nano-networks were carried in HR-TEM, (TecnaiTM G2TF20) working at an accelerating voltage of 200 kV.
Samples preparations methods for various characterizations
The prepared Rh nano-networks have been characterized using different analytical techniques such as XRD, UV-Vis, HR-TEM, EDS and Colour mapping analysis. The colloidal solution of Rh- nano-networks was directly analysed for the UV-Vis analysis. For HR-TEM analysis, the samples were prepared by drop-casting the colloidal solution of Rh nano-networks onto a Cu-coated carbon grid and proceed for slow evaporation at ambient conditions. Further, the characterizations such as XRD analyses was analysed using glass slides as substrate for the thin film preparation. The slides were initially processed with acetone and sonicated for 30 minutes. With the cleaned surface of the glass slides, the colloidal solution of Rh nano-networks has been coated by drop-casting multiple times to form thick layers. The prepared glass slides were used for XRD analysis. For SERS application, different concentrated methylene blue (MB) was prepared using DI water. The mixture of Rh nano-networks and the MB solution was coated over glass slide and dried at room temperature. The dried glass slides were further preceded for SERS analysis.
Synthesis of Rh nano-networks on bio-molecular DNA scaffolds
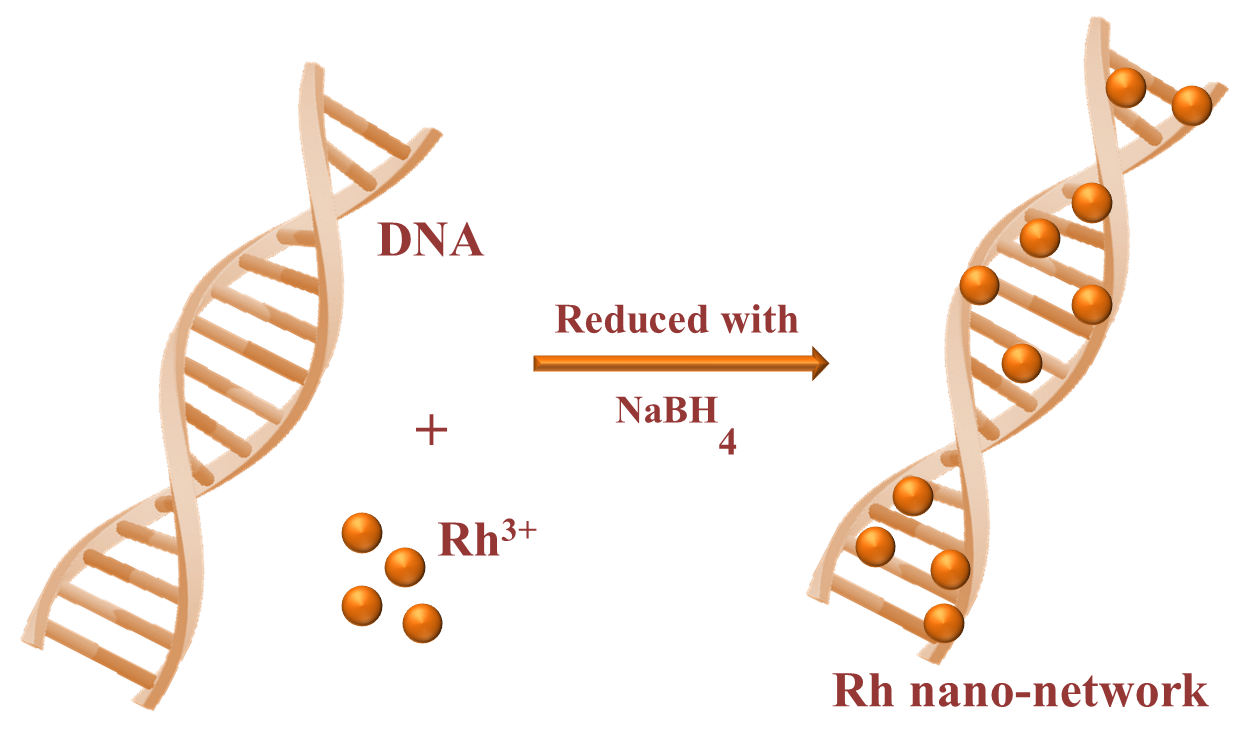
The designing of Rh nano-networks has been carried by simple wet-chemical method as given below in Figure 1. The DNA stock solution was prepared preciously by continuous stirring of DNA powder in Milli-Q water for 12 hours to get a clear solution (0.12 M).\
Figure 1. Synthetic method of forming Rh nanonetworkson DNA.
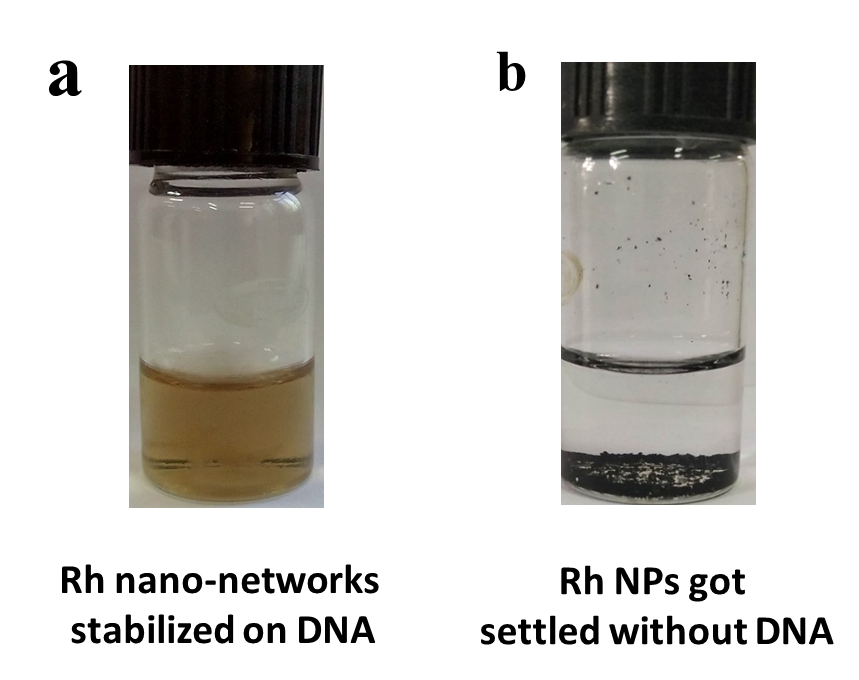
Initially, 1 ml of 0.1 M RhCl3.H2O was mixed with 4 ml of DNA stock solution and kept at stirring for 20 minutes to achieve the electrostatic interaction between Rh3+ and DNA solution. Further, the freshly prepared ice-cold solution of 0.1 M NaBH4 was added to the above mixture and the stirring continued for another 20 minutes. The orange colour appeared initially was completely turned to light brown at the end of the reaction. The observed brown colour clearly indicates the formation of Rh NPs on DNA as nano-networks. Here, the DNA plays a crucial role of stabilizing the Rh NPs in the aqueous medium and it is stable for more than six months while keeping in refrigerator. In case of without DNA, following the same method of preparation, we have tried to stabilized Rh NPs in aqueous medium. Without the presence of DNA, the absence of electrostatic interaction makes the Rh NPs unstable in the aqueous solution. This can be explained more clearly with the photographic image of Rh NPs stabilized on DNA (Figure 2a) and the settled NPs observed in the absence of DNA (Figure 2b). The immediate settle down of Rh NPs was observed, thus it implies the importance of DNA in the preparation of stable colloidal solution. The prepared Rh nano-networks were proceeded for various characterizations.
Figure 2. Photographic image of Rh NPs stabilized with DNA (a) and without DNA (b).
Results and Discussion
UV-Visible analysis
The as prepared colloidal suspension of Rh nanonetworks was analyzed by X-ray diffraction (XRD) studies as given in our previous reports [25]. Because of the particle size are too small to diffract X-ray, we could not spot any strong diffraction pattern for Rh nano-networks. Only a broad hump was observed for glass substrate, which is used to prepare sample.
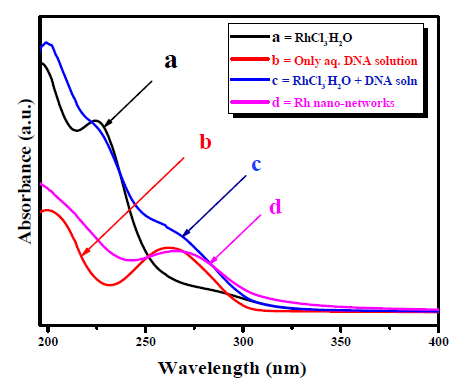
The results from the XRD clarifies that the prepared Rh nano-networks were ultra-small in size. The same kind of XRD pattern was observed in our previous studies such as the preparation of Pt, Ir, Re and RuO2 nanoparticles [11,15,39]. Further, we used to study the interaction of DNA with the metal ions in the Rh nano-networks formation using UV-Vis spectroscopy (UV-Vis) (Figure 3). At first, the absorption spectra of aqueous Rh3+ shows peak at 224.7 nm corresponds to ligand to metal charge transfer (LMCT) occurs at spin complex (curve a) of [RhCl3(H2O)]. For only DNA, the strong absorption peak at 262 nm corresponds to π-π* transitions of aromatic functionalities present in DNA (curve b). In case of mixing both aqueous RhCl 3 and DNA solution given in curve c shows shift in the absorption region from 224.7 nm to 230 nm clearly showing the interaction and nucleation of metal ions with DNA chain. Also, in curve c the presence of DNA absorption peak with the shift was identified.
Figure 3. UV-Vis absorption study of Rh nanonetworks on DNA: Curve a is the absorption peak for aqueous Rh3+ solution; Curve b is the absorption peak for aqueous DNA solution; Curve c is the absorption peak for the mixture of DNA and the Rh3+ solution and Curve d is the absorption peak for the prepared Rh nano-networks.
Moreover, the prepared Rh nano-networks were analyzed and shows the strong peaks at 268 nm was further evidenced the strong interaction of DNA with the metal ion (curve d) [25]. This kind of interaction was studied in detail previously [11,15,31].
Morphological and elemental analysis of the Rh NPs
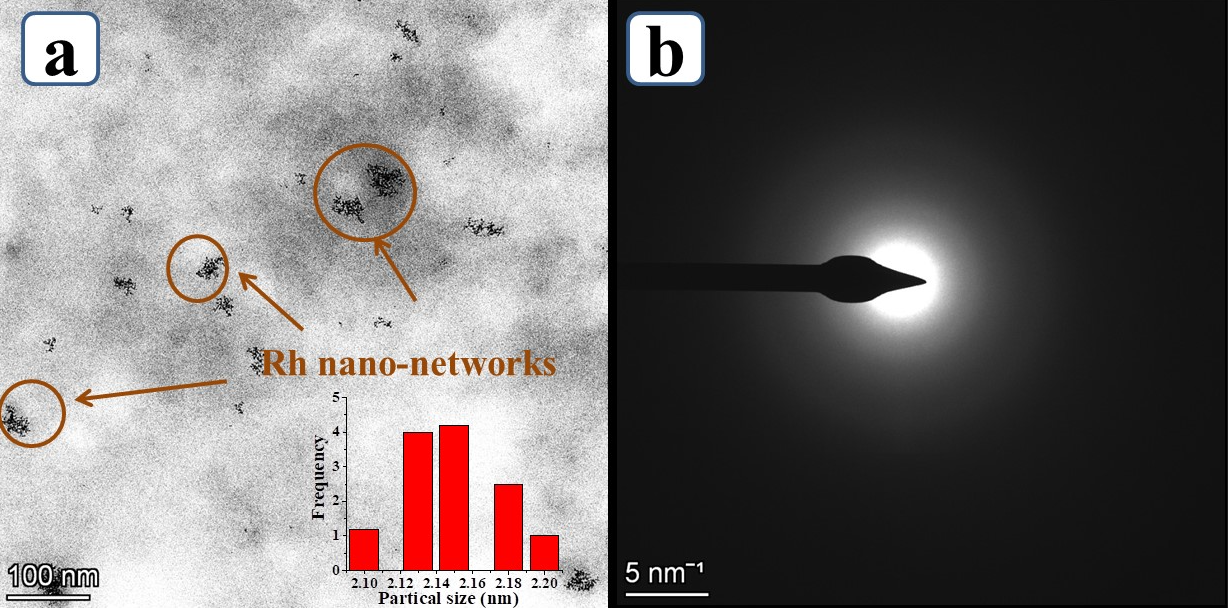
After the interaction study, the structural analysis was carried by using high resolution transmission electron microscope (HR-TEM) for Rh NPs on DNA. From the high magnified image, we could clearly see the Rh NPs were formed over DNA as nano-networks were shown in Figure 4a. We observed that the particles size of the Rh NPs as nano-networks was below 5 nm and the chain length was found to be ~98 nm. Also, the inset histogram in Figure 4a shows the average particle size calculated were 2.2 ± 0.5 nm for Rh nano-networks. Figure 4b shows the selected area electron diffraction (SAED) for Rh nano-networks gives only diffused ring. The reason behind this diffused ring was the sub-nano size of the Rh nano-networks which could not be able to diffract the electron beams. The collective results from the HR-TEM analysis portraits the deep interaction and the stable nano-networks formation of Rh NPs on DNA.
Figure 4. (a) High magnified HR-TEM analysis of Rh nano-networks and (b) corresponds to selected area electrondiffraction (SAED) for Rh nano-networks.
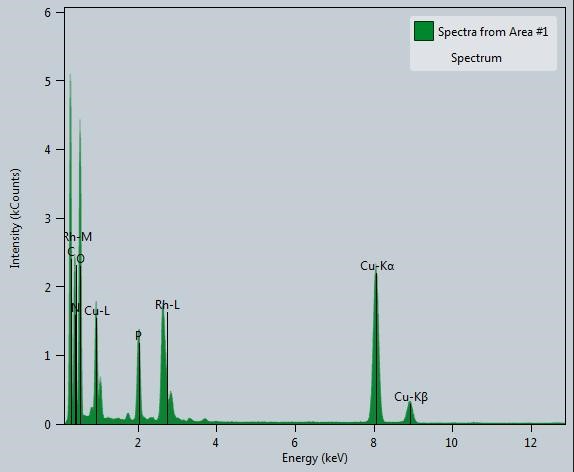
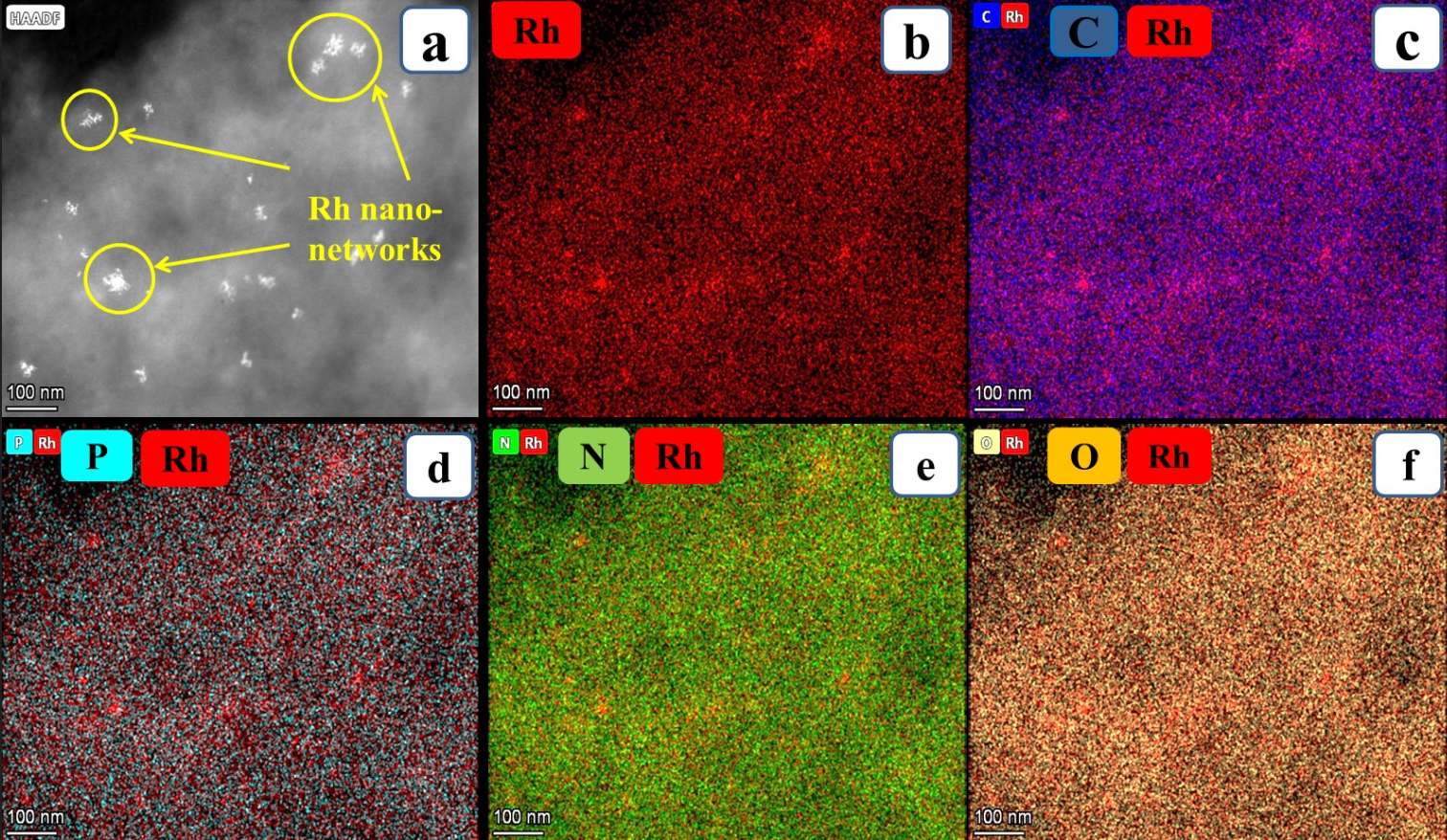
Further to identify the elemental presence, energy dispersive X-ray spectroscopy (EDS) from HR-TEM (Figure 5) has been done. The spectrum shows the presence of Rh, C, N, O, P and Cu for the Rh nano-networks. The presence of C, O, N and P are mainly from the aromatic base pairs, sugar moieties and phosphate groups from DNA. From Figure 5, the presence of Rh along with N, P, C and O further ensures that the as formed Rh NPs were congregated over DNA and gives self-assembled structures. In addition to the electronic and microstructural studies, EDS also confirms the presence of Rh NPs with DNA. The spectrum does not show any extra peaks which clearly shows the purity of the same and the copper observed was due to the copper grid used for HR-TEM analysis. Similar kind of information was obtained from the colour mapping carried in HR-TEM, the presence of Rh, C, P, N and O were present and evenly distributed over the nano-networks (Figure 6). Especially, the presence of Rh is particularized more in the marked area of HR-TEM image (Figure 6a), could suggest the gathering of Rh over DNA (Figure 6b). Also, the presence of elements in DNA conjugated with Rh was shown in Figure 6c-f. Overall from the characterization results, the Rh NPs formed on DNA were highly pure.
Figure 5. Energy dispersive X-ray spectroscopy (EDS) of the Rh nano-networks.
Figure 6. The low magnified HR-TEM images of Rh nano-networks shows the elemental mapping of Rh nanonetworks which contains the presence of Rh, C, P, N and O (a and b); The individual elemental mapping of Rh conjugated with elements present in DNA such as C, P, N and O (c-f).
SERS applications of Rh nano-networks
After the discovery of SERS, the detection limit increased up to a single-molecule level. Two theories supporting the enhancement behaviour of these Raman signals, and one is Electromagnetic field theory (EMFT) and Chemical theory (CT). In both cases, the enhancement with this Raman signal was purely depends on the localized surface plasmon resonance (LSPR) behaviour of the noble metals. But still, there is no precise cut mechanism that was proposed for this enhancement behaviour. In the Electromagnetic field theory (EMFT), the incident laser light interacts with LSPR of the plasmonic noble metals. It generates a secondary electric field, thus increases the enhancement in the EMF of the NMs. Among these two theories, the contribution of signal enhancement mainly depends on EMFT (104 to 107 orders of enhancement factor (EF)) [18]. Another critical factor is that the analyte molecules should strongly adsorb on the nanomaterial with minimal space concerning the particles. Thus, the closer interparticle distance profits the increased plasmonic resonance (10 nm) [40]. Another mechanism is that Chemical theory (CT), here the enhancement in the Raman signal purely depends on the charge transfer mechanism of the analyte molecules, and the NMs form a chemical bond between them. The effect and the probability of Raman signal enhancement are comparatively less than EMFT.
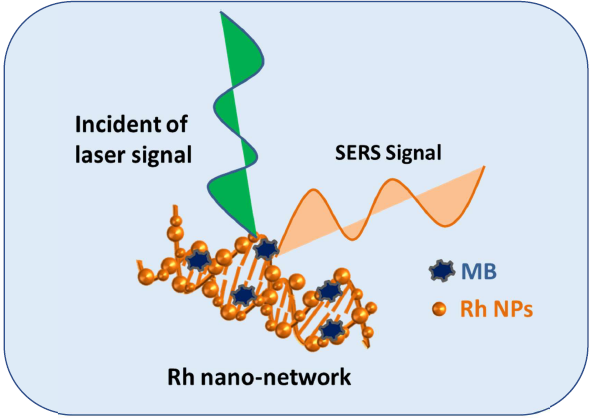
The most considered SERS substrates are Au and Ag with high enhancement factors and some other reports such as Re, Pt, and Os were studied in detail. There are very few reports when considering Rh as a SERS substrate for probe detection. In our studies, the Rh nano-networks were developed on DNA forms nano-networks with the interparticle gap of below 5 nm (Scheme 1). The nature of DNA with their double-helix structure offers a close pack network arrangement of Rh NPs over DNA (Figure 4). Thus, the reduced interparticle gap creates more number of hot spots concurrently increases the SERS signal. This cause an increased EM effect with increased EF. This kind of enhanced signal with reduced interparticle gap using DNA was detailed by our group for the preparation of Ag NPs [ 9]. The probe molecule we have chosen here is Methylene Blue (MB), which is more suitable as it fully soluble in aqueous medium, low cost and gives clear distinct peaks between 400 and 2000 cm-1. At first, we prepared various concentrated MBs from 10-3, 10-4, 10-5 and diluted up to 10-6 M. In SERS analysis, 100 μl of MB and 100 μl of Rh nano-networks were mixed together and sonicated for 5 minutes. Then the mixture was drop-casted over 1 × 1 cm glass plate and dried for over-night. The dried glass plate was preceded for SERS analysis. The SERS observed for MB with the peak values 445, 1391 and 1620 cm-1. The MB detection was possible even with the dilute concentration of 10-6 M with Rh nano-networks and the highest EF measured with the peak intensity appeared for MB was calculated to be 1.19 × 105. The SERS enhancement factor (EF) is used determine the enhanced Raman intensity is written as [7].
Scheme 1: SERS study taking Rh networks on DNA as substrate and MB as a probe molecule.
The trials made without any Rh nano networks, shows no peak for MB clearly picturize the efficiency of the developed SERS substrate using biomolecule DNA. Also, the durability of the Rh nano-networks was verified after six months of keeping in the as-synthesized solution in a refrigerator and observed the same EF with the probe detection of MB. We believe this kind of bio-molecule based noble metal NMs will be a capable player in biomedical applications.
Conclusion
In this commentary, we have highlighted the fast synthesis of ultra-small Rh nano-networks by a simple wet chemical method. The DNA used here plays a dual role in stabilizing the Rh NPs in the aqueous medium and helps to grow chain-like network morphology and also to reduce the NMs interparticle gap below 5 nm which is highly useful for generation of ‘hot spots’ in SERS studies. The characterization results portrait the strong interaction of Rh NPs with the bio-molecule DNA forms nano-networks with an average length of ~20 nm. The SERS efficiency of Rh nano-networks was screened using methylene blue (MB) as a probe molecule. The highest enhancement factor (EF) calculated was 1.19 × 105 for the lowest probe detection limit of 10-6 M was achieved with Rh nano-networks. In the future, the bio-mediated NMs will be effectively used as a SERS substrate in the bio-medical field, particularly in the cancer cell detection.
References
2. Fleischmann M, Hendra P, McQuillan A. RAMAN SPECTRA OF PYRIDINE ADSORBED AT A SILVER ELEC. Chemical physics letters. 1974 May 15;26(2).
3. Jeanmaire DL, Vanduyne RP. Heterocyclic, Aromatic, And Aliphatic Amines Adsorbed On The Anodized Silver Electrode Journal of Electroanalytical Chemistry and Interfacial Electrochemistry. 84: 1 doi: 10.1016. S0022- 0728 (77). 1977:80224-6.
4. Kundu S. Formation of self-assembled Ag nanoparticles on DNA chains with enhanced catalytic activity. Physical Chemistry Chemical Physics. 2013;15(33):14107-19.
5. Li Y, Qi X, Lei C, Yue Q, Zhang S. Simultaneous SERS detection and imaging of two biomarkers on the cancer cell surface by self-assembly of branched DNA–gold nanoaggregates. Chemical Communications. 2014;50(69):9907-9.
6. Ellington AD, Szostak JW. Selection in vitro of singlestranded DNA molecules that fold into specific ligandbinding structures. Nature. 1992 Feb;355(6363):850-2.
7. Mehn D, Morasso C, Vanna R, Bedoni M, Prosperi D, Gramatica F. Immobilised gold nanostars in a paper-based test system for surface-enhanced Raman spectroscopy. Vibrational Spectroscopy. 2013 Sep 1;68:45-50.
8. Camden JP, Dieringer JA, Wang Y, Masiello DJ, Marks LD, Schatz GC, Van Duyne RP. Probing the structure of single-molecule surface-enhanced Raman scattering hot spots. Journal of the American Chemical Society. 2008 Sep 24;130(38):12616-7.
9. Majumdar D, Singha A, Mondal PK, Kundu S. DNAmediated wirelike clusters of silver nanoparticles: an ultrasensitive SERS substrate. ACS applied materials & interfaces. 2013 Aug 28;5(16):7798-807.
10. Mandal M, Jana NR, Kundu S, Ghosh SK, Panigrahi M, Pal T. Synthesis of Au core–Ag shell type bimetallic nanoparticles for single molecule detection in solution by SERS method. Journal of Nanoparticle Research. 2004 Feb 1;6(1):53-61.
11. Sankar SS, Sangeetha K, Karthick K, Anantharaj S, Ede SR, Kundu S. Pt nanoparticle tethered DNA assemblies for enhanced catalysis and SERS applications. New Journal of Chemistry. 2018;42(19):15784-92.
12. Kundu S, Yi SI, Ma L, Chen Y, Dai W, Sinyukov AM, Liang H. Morphology dependent catalysis and surface enhanced Raman scattering (SERS) studies using Pd nanostructures in DNA, CTAB and PVA scaffolds. Dalton Transactions. 2017;46(29):9678-91.
13. Sakthikumar K, Anantharaj S, Ede SR, Karthick K, Kundu S. A highly stable rhenium organosol on a DNA scaffold for catalytic and SERS applications. Journal of Materials Chemistry C. 2016;4(26):6309-20.
14. Nithiyanantham U, Ede SR, Kundu S. Self-assembled wire-like and honeycomb-like osmium nanoclusters (NCs) in DNA with pronounced catalytic and SERS activities. Journal of Materials Chemistry C. 2014;2(19):3782-94.
15. Sakthikumar K, Anantharaj S, Ede SR, Karthick K, Ravi G, Karthik T, Kundu S. Prompt synthesis of iridium organosol on DNA for catalysis and SERS applications. Journal of Materials Chemistry C. 2017;5(45):11947-57.
16. Kumaravel S, Thiruvengetam P, Karthick K, Sankar SS, Kundu S. Detection of Lignin Motifs with RuO2-DNA as an Active Catalyst via Surface-Enhanced Raman Scattering Studies. ACS Sustainable Chemistry & Engineering. 2019 Oct 22;7(22):18463-75.
17. Su Q, Ma X, Dong J, Jiang C, Qian W. A reproducible SERS substrate based on electrostatically assisted APTESfunctionalized surface-assembly of gold nanostars. ACS applied materials & interfaces. 2011 Jun 22;3(6):1873-9.
18. Culha M, Cullum B, Lavrik N, Klutse CK. Surface-enhanced Raman scattering as an emerging characterization and detection technique. Journal of Nanotechnology. 2012 Jan 1;2012.
19. Ren B, Lin XF, Yang ZL, Liu GK, Aroca RF, Mao BW, Tian ZQ. Surface-enhanced Raman scattering in the ultraviolet spectral region: UV-SERS on rhodium and ruthenium electrodes. Journal of the American Chemical Society. 2003 Aug 13;125(32):9598-9.
20. Shen J, Su J, Yan J, Zhao B, Wang D, Wang S, Li K, Liu M, He Y, Mathur S, Fan C. Bimetallic nano-mushrooms with DNA-mediated interior nanogaps for high-efficiency SERS signal amplification. Nano Research. 2015 Mar 1;8(3):731-42.
21. McLellan JM, Siekkinen A, Chen J, Xia Y. Comparison of the surface-enhanced Raman scattering on sharp and truncated silver nanocubes. Chemical Physics Letters. 2006 Aug 18;427(1-3):122-6.
22. Caprara D, Ripanti F, Capocefalo A, Sarra A, Brasili F, Petrillo C, Fasolato C, Postorino P. DNA-functionalized gold nanoparticle assemblies for Surface Enhanced Raman Scattering. Colloids and Surfaces A: Physicochemical and Engineering Aspects. 2020 Feb 20;589:124399.
23. Wei G, Wang L, Liu Z, Song Y, Sun L, Yang T, Li Z. DNA-network-templated self-assembly of silver nanoparticles and their application in surface-enhanced Raman scattering. The Journal of Physical Chemistry B. 2005 Dec 22;109(50):23941-7.
24. Kundu S, Yi SI, Ma L, Chen Y, Dai W, Sinyukov AM, Liang H. Morphology dependent catalysis and surface enhanced Raman scattering (SERS) studies using Pd nanostructures in DNA, CTAB and PVA scaffolds. Dalton Transactions. 2017;46(29):9678-91.
25. Sangeetha K, Sankar SS, Karthick K, Anantharaj S, Ede SR, Wilson S, Kundu S. Synthesis of ultra-small Rh nanoparticles congregated over DNA for catalysis and SERS applications. Colloids and Surfaces B: Biointerfaces. 2019 Jan 1;173:249-57.
26. Chen Z, Liu C, Cao F, Ren J, Qu X. DNA metallization: principles, methods, structures, and applications. Chemical Society Reviews. 2018;47(11):4017-72.
27. Kang Z, Yan X, Zhang Y, Pan J, Shi J, Zhang X, Liu Y, et.al. Single-stranded DNA functionalized single-walled carbon nanotubes for microbiosensors via layer-by-layer electrostatic self-assembly. ACS applied materials & interfaces. 2014 Mar 26;6(6):3784-9.
28. Zhao H, Dong J, Zhou F, Li B. One facile fluorescence strategy for sensitive detection of endonuclease activity using DNA-templated copper nanoclusters as signal indicators. Sensors and Actuators B: Chemical. 2017 Jan 1;238:828-33.
29. John J, Thomas L, Kurian A, George SD. Enhanced heat diffusion in nanofluid via DNA mediated aggregation. RSC advances. 2016;6(67):62390-8.
30. Wilner OI, Willner I. Functionalized DNA nanostructures. Chemical reviews. 2012 Apr 11;112(4):2528-56.
31. Kumaravel S, Thiruvengetam P, Karthick K, Sankar SS, Kundu S. Detection of Lignin Motifs with RuO2-DNA as an Active Catalyst via Surface-Enhanced Raman Scattering Studies. ACS Sustainable Chemistry & Engineering. 2019 Oct 22;7(22):18463-75.
32. Abdel-Rahman LH, Abu-Dief AM, Basha M, Hassan Abdel-Mawgoud AA. Three novel Ni (II), VO (II) and Cr (III) mononuclear complexes encompassing potentially tridentate imine ligand: Synthesis, structural characterization, DNA interaction, antimicrobial evaluation and anticancer activity. Applied Organometallic Chemistry. 2017 Nov;31(11):e3750.
33. Abd El-Aziz DM, El-Wakiel N, Gaber M. Fluorescent UO2 (II) and ZrO (II) complexes: Synthesis, structural characterization, fluorescence, DNA binding studies and biological applications in cell probing. Applied Organometallic Chemistry. 2019 Aug;33(8):e4855.
34. Abu-Dief AM, Abdel-Rahman LH, Abdel-Mawgoud AA. A robust in vitro Anticancer, Antioxidant and Antimicrobial Agents Based on New Metal-Azomethine Chelates Incorporating Ag (I), Pd (II) and VO (II) Cations: Probing the Aspects of DNA Interaction. Applied Organometallic Chemistry. 2020 Feb;34(2):e5373.
35. Samanta A, Medintz IL. Nanoparticles and DNA–a powerful and growing functional combination in bionanotechnology. Nanoscale. 2016;8(17):9037-95.
36. Turunc E, Binzet R, Gumus I, Binzet G, Arslan H. Green synthesis of silver and palladium nanoparticles using Lithodora hispidula (Sm.) Griseb.(Boraginaceae) and application to the electrocatalytic reduction of hydrogen peroxide. Materials Chemistry and Physics. 2017 Dec 1;202:310-9.
37. Goddard G, Brown LO, Habbersett R, Brady CI, Martin JC, Graves SW, et.al. High-resolution spectral analysis of individual SERS-active nanoparticles in flow. Journal of the American Chemical Society. 2010 May 5;132(17):6081-90.
38. Wang Z, Zhang J, Ekman JM, Kenis PJ, Lu Y. DNAmediated control of metal nanoparticle shape: one-pot synthesis and cellular uptake of highly stable and functional gold nanoflowers. Nano letters. 2010 May 12;10(5):1886-91.
39. Nithiyanantham U, Ede SR, Kundu S. Self-assembled wire-like and honeycomb-like osmium nanoclusters (NCs) in DNA with pronounced catalytic and SERS activities. Journal of Materials Chemistry C. 2014;2(19):3782-94.
40. Zhang L, Ma H, Yang L. Design and fabrication of surface-enhanced Raman scattering substrate from DNA– gold nanoparticles assembly with 2–3 nm interparticle gap. RSC Advances. 2014;4(85):45207-13.