Introduction
Bacterial Toxin-Antitoxin (TA) modules were extensively studied [1-3]. They are located on extra chromosomal genetic elements as well as on bacterial chromosomes. They carry two genes. One encoding for the toxin, and the other for the antitoxin. The toxin is a stable protein, while the antitoxin is either RNA or a labile protein. They are divided into six groups [3,4], among them group II is the most studied. In-group II both toxin and antitoxin are proteins [5]. To this group belongs the E. coli TA system mazEF which is the first bacterial chromosomal TA discovered, and is since extensively investigated [6-9]. The toxin MazF is a stable protein while the antitoxin MazE is degraded by ClpX protease [6]. mazEF is triggered by all kind of stressful conditions like rifampicin, chloramphenicol, spectinomycin, high temperature (50°C) [7,10], DNA damage (UV irradiation, nalidixic acid, mitomycin C. thymine starvation) and oxidative stress (H2O2) [7,10-12]. Each of these stressful conditions has been shown to lead to E. coli mazEF-dependent cell death, which occurs only at logarithmic stage of growth in minimal medium. In addition, E. coli mazEF-mediated cell death is a population phenomenon requiring a unique E. coli quorum sensing (QS) factor, the Extracellular Death Factor (EDF) [13,14]. By structural analysis, E. coli EDF (EcEDF) was identified as the linear penta-peptide Asn-Asn-Trp-Asn-Asn (NNWNN) [13]. EcEDF amplifies both the endoribonucleolytic activities of E. coli MazF and overcomes the inhibitory activity of the E. coli antitoxin MazE over the toxin MazF [15]. E. coli MazF is a sequence-specific endoribonuclease that preferentially cleaves single-stranded mRNAs at ACA sites [16], and thereby generate a special translation machinery designated STM (Stress Translation Machinery [17]. This occurs by the following process: a) MazF creates "stress mRNAs" by cleaving at ACA sites immediately upstream to [17], or further upstream [18] from the AUG-start codons of specific mRNAs; b) by targeting an ACA site in the 16S rRNA within the 30S ribosomal subunit at the decoding center, MazF removes 43 nucleotides from the 3’-terminus, including the anti-Shine Dalgarno region [17].
Here we investigated the intracellular localization of the protein products of the following type II TA modules: mazEF, chpBIK, mqsRA and rnlAB [19]. We followed the localization of these proteins by fusing them with the fluorescent protein mCherry. Then, we used fluorescent microscopy and image analysis software in order to obtain the protein distribution data.
Our results revealed that with the exception of the chpBIK TA module, the localization of each toxin-antitoxin complex differ from the localization of the toxin by itself. Thus, our results clearly show that the antitoxin shifts the localization of its respective toxin towards the center of the cell. Probably such a shift contributes to a reduction of cellular toxicity.
Results and Discussion
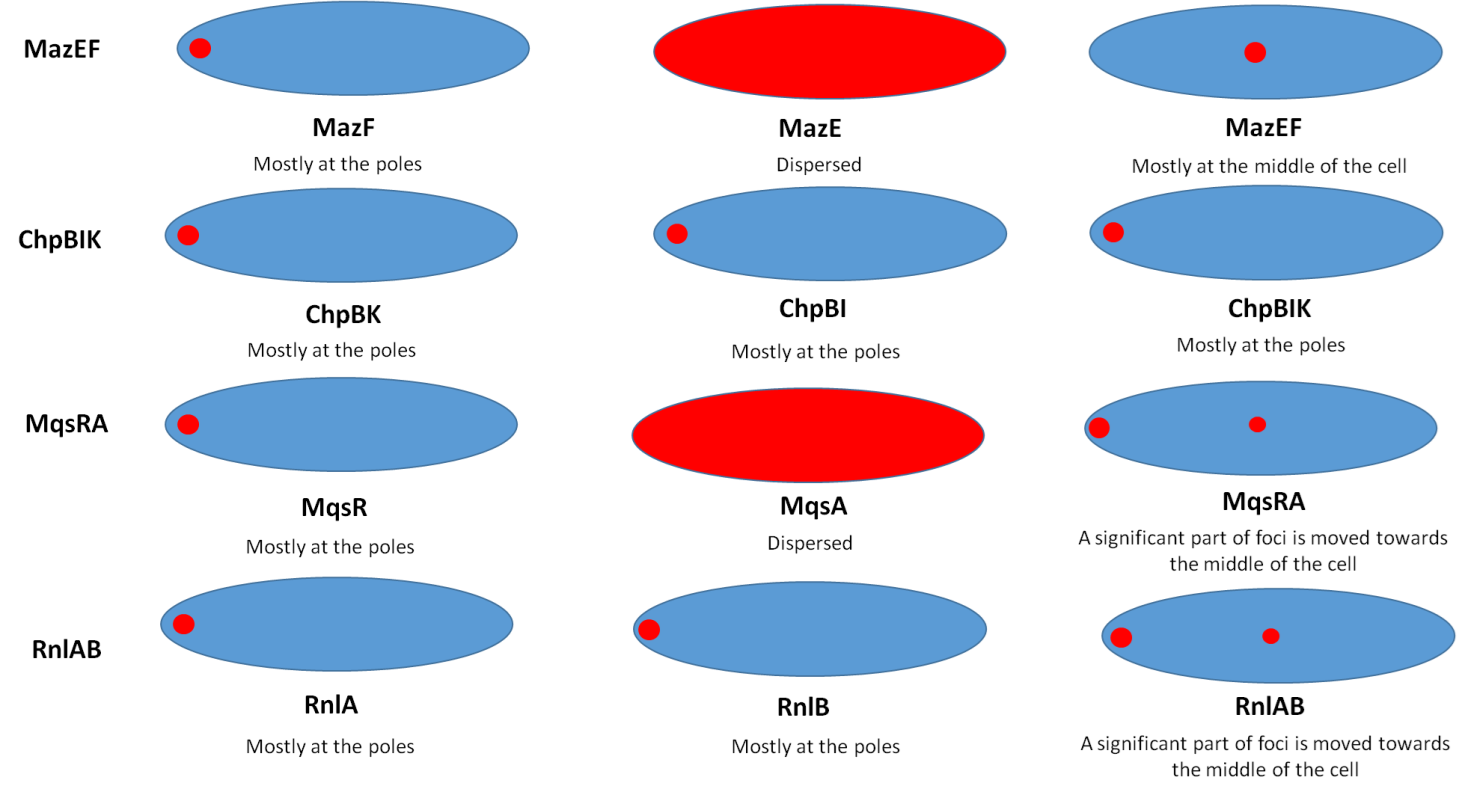
In contrast to our expectation each of the studied TA modules had its own pattern of localization for the toxin, the antitoxin, and the toxin-antitoxin complex (Figure 1). With the exception of the chpBIK TA module, for each TA system, the localization of the toxin-antitoxin complex differs the localization of the toxin by itself. The presence of the antitoxin shifts the toxin from the pole towards the center of the cells. Therefore, we suggest that in contrast to the generally considered view, the antagonistic effect of antitoxins on their cognate toxins is not only based on their direct structural interactions. But it is also caused by changing the intracellular localization of the toxin. The reasons for the migration of the toxins toward the poles, and their shifts toward the center of the cells in the presence of the antitoxins, are subjects for future experiments.
Figure 1. A schematic representation of the intracellular localization of the protein products of some type II TA systems in E. coli.
The representation was taken from the fluorescent microscopy images of the protein-mCherry fusions [19]. Fluorescence is represented by red color. Larger red circles represent polar localization (MazF and ChpBK), and smaller red circles represent the localization at the middle of the cells (MqsRA, RnlAB). Dispersed localization (MazE, MqsA) is represented as a full colored red bacterial cell.
Further studies should identify the E. coli component(s) that provide the peculiar location pattern of group II TA modules. Is it related to E. coli proteins that were previously identified as being critical for cell division [20-22]? Or to those that have other functions and were identified to have a polar localization [23-24]? Alternatively, is the polar localization connected to the yet unknown location of the STM or EDF systems that are unique at least in the case of MazF?
References
2. Fraikin N, Goormaghtigh F, Van Melderen L. Type II toxin-antitoxin systems: evolution and revolutions. Journal of Bacteriology. 2020 Mar 11;202(7):e00763-19.
3. Yamaguchi Y, Inouye M. Toxin–antitoxin systems in bacteria and archaea. Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. 2016 Aug 26:97-107.
4. Page R, Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nature Chemical Biology. 2016 Apr;12(4):208-14.
5. Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annual Review of Genetics. 2011 Dec 15;45:61-79.
6. Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal" addiction module" regulated by guanosine [corrected] 3', 5'-bispyrophosphate: a model for programmed bacterial cell death. Proceedings of the National Academy of Sciences. 1996 Jun 11;93(12):6059-63.
7. Hazan R, Sat B, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. Journal of Bacteriology. 2004 Jun 1;186(11):3663-9.
8. Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genetics. 2009 Mar 13;5(3):e1000390.
9. Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. Journal of Biological Chemistry. 2005 Feb 4;280(5):3143-50.
10. Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. Journal of Bacteriology. 2001 Mar 15;183(6):2041-5.
11. Sat B, Reches M, Engelberg-Kulka H. The Escherichia coli mazEF suicide module mediates thymineless death. Journal of Bacteriology. 2003 Mar 15;185(6):1803-7.
12. Kolodkin-Gal I, Engelberg-Kulka H. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. Journal of Bacteriology. 2006 May 1;188(9):3420-3.
13. Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science. 2007 Oct 26;318(5850):652-5.
14. Kumar S, Kolodkin-Gal I, Vesper O, Alam N, Schueler-Furman O, Moll I, Engelberg-Kulka H. Escherichia coli quorum-sensing EDF, a peptide generated by novel multiple distinct mechanisms and regulated by trans-translation. MBio. 2016 Jan 26;7(1):1-12.
15. Belitsky M, Avshalom H, Erental A, Yelin I, Kumar S, London N, et al. The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Molecular Cell. 2011 Mar 18;41(6):625-35.
16. Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. Journal of Biological Chemistry. 2005 Feb 4;280(5):3143-50.
17. Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011 Sep 30;147(1):147-57.
18. Sauert M, Wolfinger MT, Vesper O, Müller C, Byrgazov K, Moll I. The MazF-regulon: a toolbox for the post-transcriptional stress response in Escherichia coli. Nucleic Acids Research. 2016 Aug 19;44(14):6660-75.
19. Mager A, Safran T, Engelberg-Kulka H. Intracellular Localization of the Proteins Encoded by Some Type II Toxin-Antitoxin Systems in Escherichia coli. Mbio. 2021 Aug 3;12(4):1-15.
20. Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proceedings of the National Academy of Sciences. 1996 Nov 12;93(23):12998-3003.
21. Romberg L, Levin PA. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annual Reviews in Microbiology. 2003 Oct;57(1):125-54.
22. Masuda H, Tan Q, Awano N, Yamaguchi Y, Inouye M. A novel membrane-bound toxin for cell division, CptA (YgfX), inhibits polymerization of cytoskeleton proteins, FtsZ and MreB, in Escherichia coli. FEMS Microbiology Letters. 2012 Mar 1;328(2):174-81.
23. Lybarger SR, Maddock JR. Polarity in action: asymmetric protein localization in bacteria. Journal of Bacteriology. 2001 Jun 1;183(11):3261-7.
24. Lopian L, Elisha Y, Nussbaum-Shochat A, Amster-Choder O. Spatial and temporal organization of the E. coli PTS components. The EMBO Journal. 2010 Nov 3;29(21):3630-45.