Abstract
Background: Fluoroquinolone antibiotics are usually used for the treatment of urinary tract infections. The aim of this study was to determine the prevalence and molecular characterization of Plasmid-Mediated Quinolone Resistance (PMQR) genes among ESBL-producing Escherichia coli isolates obtained from tertiary referral hospital in Tehran, Iran.
Methods: A total of 150 uropathogenic E. coli isolates were obtained from Rasool-e- Akram hospital. The bacterial isolates were identified by standard laboratory methods. Then, the susceptibility to quinolone antibiotics was assessed by standard disk diffusion method. The PCR method was used to show presence of qnrA, qnrB, qnrS, aac(6)-Ibcr and qepA genes.
Results: Overall, 79 of the 150 isolates (52.6%) were non-susceptible to quinolone antibiotics. Out of 79 quinolone non-susceptible isolates, 46 (58.2%) isolates harboured PMQR-encoding genes. Further, 36 (24%) had aac(6)-Ib-cr gene and interestingly, amplification assays showed that 33 (41.8%) out of 79 quinolone non-susceptible isolates carried only qnrB gene. Also qnrA, qnrB (30.9%), qepA (7.3%) and qnrS (25.4%) genes showed.
Conclusions: This study showed a high prevalence of aac(6)-Ib-cr, qnrB, qnrS and qnrA genes in the uropathogenic E. coli isolates from tertiary referral hospital. Therefore, the application of proper infection control and well-established antibiotic prescription guidelines seems to be highly needed in our medical centers.
Keywords
Quinolones resistance, Urinary tract infection, aac(6)-Ib-cr, Pyelonephritis, Cystitis
Introduction
Urinary Tract Infection (UTI) due to Escherichia coli is the most common bacterial infection. Fluoroquinolones are commonly used for the treatment of UTI because isolated microorganisms are frequently resistant to aminopenicillins and trimethoprim- sulfamethoxazole and fluoroquinolones are given orally [1].
Quinolones are among the main groups of antimicrobial compounds which are used against bacterial infections. However, many recent studies have shed light on the fact that the resistance of various members of Enterobacteriaceae, likely Salmonella and Klebsiella, to quinolones and fluoroquinolones is increasing, globally [2]. Resistance to the new generations of antibiotics and thereafter the development of resistant strains has become prevalent not only among the nosocomial infection but also among community-acquired infections.
Recent studies have highlighted the increasing rate of the detection of quinolone-resistant E. coli strains [3]. Interestingly, the infections by such strains show a multi-drug resistance to beta-lactams and aminoglycosides [4]. Recent founding showed that the Plasmid-Mediated Quinolone Resistance (PMQR) may be the main cause of bacterial resistance to quinolones. There are three major groups of gene markers on qnr plasmids; qnrA, qnrB, and qnrS genes [5,6].
There have never been any reports concerning the prevalence of qnr genes in E. coli isolates from patients with urinary infection in Tehran Hospitals. Thus, the aim of survey was studying the qnr markers in E. coli isolates which their ESBL capability was already confirmed. These isolates were originally isolated from children with urinary infection.
Materials and Methods
Patients
The inclusion criteria were age between 1 day until 5 years and the presence of cystitis symptoms (pain on urination, pollakiuria, supra-pubic pain). Patients with recurrent UTIs (three episodes of UTI during the common year), could be included.
Sampling urine and microbiological evaluation
Exclusion criteria were antibiotic treatment in the previous two weeks. Every patient gave her informed consent. Afresh midstream urine or catheter sample or supra pubic was collected from all eligible patients, respecting aseptic conditions. The microbiological de?nition of UTI was leukocyturia ? 10 leukocytes/mL and bacteriuria ? 105 cfu/ml in urine collected from midstream and In urine collected with catheter>102 cfu/ml for supra- pubic.
Isolation and bacterial identification of bacteria and assessment of their antibiotic susceptibility
E. coli isolates of urine samples were incubated in LB medium for 18h, 37°C. The plasmid DNA was extracted using a Bioneer kit (Kit number: K-3030-2). The used primers for the amplification of the given qnr genes are listed in (Table 1). The PCR was directed in a thermal cycler(SENSOQUEST, Germany) and the program was as below: a cycle of 94°C, 5 min followed by 35 cycles of 95°C, 1 min (denaturation), 54°C, 1 min (annealing), and 72°C, 1 min (extension). The PCR products were electrophoresed on a 2% agarose gel to visualize the amplified genomic fragments. Finally, the collected data was analyzed statistically using SPSS22 package. Results are for analyses of virulence genes as detected by PCR only.
| Target genes | PCR product | Primer sequences (5'->3') | Annealing temperatures(°C) |
|---|---|---|---|
| qnrA-F | 516(bp) | ATTTCTCACGCCAGGATTTG | 56 |
| qnrA R | GATCGGCAAAGGTTAGGTCA | ||
| qnrB-F | 469(bp) | GATCGTGAAAGCCAGAAAGG | 55 |
| qnrB R | ACGATGCCTGGTAGTTGTCC | ||
| qnrS-F | 417(bp) | ACGACATTCGTCAACTGCA | 57 |
| qnrS R | TAAATTGGCCTGTAGGC |
Results
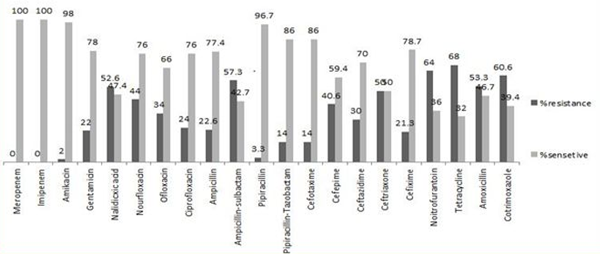
In general, 33.3% (50 people) of the studied people were outpatients and the remaining 66.6% (100 people) were in-patients. Studying these patients for 12 months, 74 (49.3%) patients showed at least a relapse of urinary infection, and in these cases, 68% of the isolates were resistant to ampicillin, (64%) tetracyclin, (60.6%) cotrimoxazole, (57.3%) piperacillin, respectively. Besides, the highest antibiotic susceptibilities were detected for Imipeneme, Meropeneme (100%), Amikacin (98%), and (96.7%) piperacillin/tazobactam, respectively (Figure 1).
Further, 82 isolates (54.6%) showed resistance to three or more antibiotic classes and were categorized as Multidrug Resistance (MDR) strains. The prevalence of antibiotic resistance genes and their distribution is demonstrated in (Tables 2).
| No (%). of positive isolates in UTI patients | Total No (%). of positive isolates (n=150) | |||
|---|---|---|---|---|
| Resistance genes | In patients (n=100) |
Out patients (n=50) | P Value | |
| qepA | 9 (9%) | 2 (4%) | 0.26 | 11 (7.3%) |
| qnrA | 4 (4%) | 1 (2%) | 0.52 | 5 (3.3%) |
| qnrB | 22 (22%) | 11 (22%) | 0.05 | 33 (22%) |
| qnrS | 18 (18%) | 9 (18%) | 0.5 | 27 (18%) |
| aac(6’)-Ib-cr | 20 (20%) | 16 (32%) | 0.105 | 36 (24%) |
Discussion
In this study, 52.6% and 24% resistance was detected to nalidixic acid and ciprofloxacin; these statistics seem to be similar to those of Peimani et al., [7]. But, our results show higher resistance prevalence to those of Raei et al., [8]. [18.57% (NA) and 36.2% (Cp)] and Zamani et al., [9].[23.8% (NA) and 34.1% (Cp)]. Considering the increasing rise of resistance to extended-spectrum antibiotics in Hospitals and also communities, an outbreak of quinolone resistant isolates in hospitals and clinical settings seem inevitable. Also, taking non-prescribed antibiotics worst the situation [7-9]. These studies highlight the particularity of ?uoroquinolone resistance in E. coli for Iran’s tertiary hospitals; however, a shortfall exists in information about the molecular epidemiology in those of county status. Also 52.6% of E. coli isolates were non-susceptible to quinolone antibiotics .Resistance to fluoroquinolones in E. coli is quite high in many European countries ranging from 25% to 50 [10].
In this study, qnrA (18%), qnrB (30.9%) and qnrS (25.4%) were detected in quinolone resistant E. coli. This report is the first which concerned the prevalence of quinolone resistance in Rasool-e-Akram Hospital, Iran. Pakzad et al., showed that 37.5% and 20.8% of the detected ESBL E. coli were qnrA+ and qnrB+, respectively [11] Saiadpour et al. showed that 30.4% of the community which they studied had qnr and/or aac(6’)-Ib-cr genes [12]. Wang et al. reported 2.4%, 6.1% and 15.1% prevalence of qnrA, qnrB and qnrS genes in the Klebsiella isolates, respectively [13]. Also, the coincidence of the qnr and bla (beta-lactamases), which is of a considerable significance, has been reported. Also, qnrA gene was detected in only five E. coli isolates. Founding of Peimani et al., [7] who haven’t detected aac(6)- Ib-cr gene in Klebsiella isolates of 250 urinary infection samples. Peimani et al. [7] also reported qnrB gene as the most prevalent gene in those Klebsiella isolates.
In this study, 46% of the qnr-positive (qnr+) isolates showed the highest resistance to quinolones. Interestingly, a survey in the ICU and CCU hospital units showed that qnr genes were detected in the isolates of the patients who experienced multiple relapses of urinary infection, annually. Also in this study qnrS, qnrB, aac(6)-Ib-cr genes were found as the most prevalent antibiotic genes in UPEC isolates. The critical role of these genes in antibiotic resistance of such bacteria has been already highlighted. Comparing the results to those of similar investigations in Iran and other countries, aac(6)-Ib-cr gene was found as the most prevalent antibiotic resistance gene in UPEC isolates. Ma J et al. [14] showed a 18.8% prevalence of aac(6)-Ib-cr gene . In Comparison, Arabi et al. [15] and Momtaz H et al. [16] indicated 34% and 46.34% prevalence of this gene in UPEC isolates. Interestingly, it has been already shown that this gene, which is located on a plasmid, is not transmitted through the wellknown plasmid transmission ways. In addition, there is a report concerning the plasmids which may have a role in the distribution of this gene. However, this gene is an Integron Cassette Insertion (ICI) indeed, and so, it might be transmitted among various plasmids. Large-scale administration of quinolones and/or cephalosporines to food-production animals might select for cephalosporinresistant (blaCTX-M) and plasmid-mediated quinolone resistant E. coli strains in animals.
The aac(6’)-Ib gene encodes a common aminoglycoside acetyltransferase responsible for resistance to aminoglycoside antibiotics such as kanamycin, amikacin and tobramycin [9]. The co transmission of qnr with aac(6’)- Ib-cr genes which speeds up the formation of multidrug resistance in Enterobacteriaceae has been previously reported in China [14]. However, in the present study we showed that currently there is no significant relationship between aac(6’)-Ib-cr prevalence and the presence of the qepA, qnrB, qnrA ,qnrS gene in Iran. According to the fact that gentamycin is being prescribed much more than that of quinolones for urinary infection patients, so it may lead to a natural selection.
In 70.1% of all isolates two or more genes responsible for ?-lactam or quinolone resistance were identified. Combinations of several ?-lactamase genes are commonly observed regularly within genomes of Gram-negative bacterial pathogens and co-selection of ?-lactamase genes and determinants for resistance against other antibiotics e.g. fluoroquinolones has been postulated as a possible mechanism responsible for the widespread distribution of those combined genes [17]. The frequently observed combination of bla CTX-M-15 and aac(6’)-Ib-cr could provide genetic support for this theory, which has been noted before [18]. However, a strong association between resistance to TMS and the presence of integrons was observed [19]. Trimethoprim-sulfamethoxazole and fluoroquinolones tend to more effective than many Betalactams, in eradicating initial bacteriuria. As a result use of beta-lactams class for only may yield increased incidence of recurrence and side effects.
Interestingly, in this work, a strong association between fluoroquinolone-resistant E. coli and qnrB was observed (p value<0 href="#17" title="17">17]. These results suggest that fluoroquinolone-susceptible E. coli strains would have more virulence determinants since they belong to group’s commensal. In contrast, there was a strong association between fluoroquinolone-resistance strains and commensal groups, suggesting that the presence of these resistance mechanisms would favor E. coli clones to become successful commensals. Interestingly, qnrB has been detected on the CTX-M-15 and SHV-12 ESBL bla coding plasmids in India and USA, respectively. Considering the fact that plasmid-born qnrB attach to the upstream of LexA [20], it can cause an increase of qnrB expression in ESBL+ E. coli when expose to quinolones. Therefore, in this study 30.9% of the isolates were qnrB+ and Peimani reported a 35.5% prevalence of this gene (7).
In this study a high prevalence of PMQR was detected in ESBL+ E. coli. Shams et al. [21] showed that their E. coli isolates were all qnrA+ and also 80% of their E. coli isolates showed a MDR characteristic. However Peimani [7] (Iran), and Bouchakour [22] (Morocco) detected 39.5%, and 50% qnrA+ E .coli. The difference of the results of this study with the above mentioned results may be related to the increase of PMQR in Enterobacteriaceae, especially E. coli [22,7].
MDR is one of the major worries of medical authorities and communities. In this study, a high prevalence of MDR in UPEC isolates was detected and 82 isolates were detected as MDR. There are various reports on the prevalence of MDR isolates. For example; 42% is reported in Slovenia and 65.6% in Iran [23]. Our results showed that there might be an increasing rate of antibiotic resistance and so an increasing rate of MDR in UPEC isolates of Hospitals and clinical settings of Tehran seems to be inevitable.
Conclusion
This study showed a high prevalence of MDQR which is related to the presence of qnr and also aac(6’)-Ib-cr genes in the E. coli isolates. The presence of such an antibiotic resistance in clinical settings of the country seems to be alarming, not only for human health, but also financially. It is particularly important to understand the prevalence and characteristics of ?uoroquinolone resistance in county hospitals so that guidance can be provided on the clinical application of these antibiotics. Our results show that more studies is needed to control the infections. Also, prevention of taking non-prescribed antibiotics is highly needed to pave the way to control such infections.
Acknowledgment
The authors thank all the participants in this study. This research has been financially supported by research center of pediatric infectious disease of Rasool-e-Akram hospital grant No: 241879.
Ethics approval
This study was approved by the institutional review board (IRB) of the Iran University of Medical Sciences, Rasool-e- Akram Hospital (IRB No. IR.IUMS.REC.1393.24879).
References
2. Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clinical microbiology reviews. 2009 Oct 1; 22(4):664-89.
3. Baquero F, Tedim AP, Coque TM. Antibiotic resistance shaping multi-level population biology of bacteria. The multiple roles of antibiotics and antibiotic resistance in nature. 2015: 24.
4. Rabasa AI, Shattima D. Urinary tract infection in severely malnourished children at the University of Maiduguri Teaching Hospital. Journal of tropical pediatrics. 2002; 48(6):359-61.
5. Fang H, Huang H, Shi Y, Hedin G, Nord CE, Ullberg M. Prevalence of qnr determinants among extended- spectrum ß-lactamase-positive Enterobacteriaceae clinical isolates in southern Stockholm, Sweden. International journal of antimicrobial agents. 2009; 34(3):268-70.
6. Yamane K, Wachino J-i, Suzuki S, Kimura K, Shibata N, Kato H, et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrobial agents and chemotherapy. 2007 Sep 1; 51(9):3354-60.
7. Peymani A, Farivar TN, Nikooei L, Najafipour R, Javadi A, Pahlevan AA. Emergence of plasmid- mediated quinolone-resistant determinants in Klebsiella pneumoniae isolates from Tehran and Qazvin provinces, Iran. Journal of preventive medicine and hygiene. 2015 Jun; 56(2):E61.
8. Raei F, Eftekhar F, Feizabadi MM. Prevalence of quinolone resistance among extended-spectrum ß-lactamase producing uropathogenic Klebsiella pneumoniae. Jundishapur journal of microbiology. 2014 Jun; 7(6).
9. Zamani A, Mashouf RY, Namvar AM, Alikhani MY. Detection of magA Gene in Klebsiella spp. Isolated from clinical samplesdetection of magA. Iranian journal of basic medical sciences. 2013 Feb; 16(2):173.
10. Iabadene H, Messai Y, Ammari H, Ramdani- Bouguessa N, Lounes S, Bakour R, Arlet G. Dissemination of ESBL and Qnr determinants in Enterobacter cloacae in Algeria. Journal of antimicrobial chemotherapy. 2008 Apr 1; 62(1):133-6.
11. Jorgensen JH, Turnidge JD. Susceptibility test methods: dilution and disk diffusion methods. In Manual of Clinical Microbiology, Eleventh Edition 2015 Jun 1 (pp. 1253-1273).
12. Pakzad I, Ghafourian S, Taherikalani M, Abtahi H, Rahbar M, Mansory Jamshidi N. Qnr prevalence in extended spectrum beta-lactamases (ESBLs) and none- ESBLs producing Escherichia coli isolated from urinary tract infections in central of Iran. Iranian journal of basic medical sciences. 2011; 14(5):458-64.
13. Seyedpour SM, Eftekhar F. Quinolone Susceptibility and Detection of qnr and aac (6’)-Ib-cr Genes in Community Isolates of Klebsiella pneumoniae. Jundishapur journal of microbiology. 2014 Jul; 7(7).
14. Wang M, Jacoby GA, Mills DM, Hooper DC. SOS regulation of qnrB expression. Antimicrobial agents and chemotherapy. 2009 Feb 1; 53(2):821-3.
15. Ma J, Zeng Z, Chen Z, Xu X, Wang X, Deng Y, Lü D, Huang L, Zhang Y, Liu J, Wang M. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac (6')-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrobial agents and chemotherapy. 2009 Feb 1; 53(2):519-24.
16. Arabi S, Tohidi F, Naderi S, Nazemi A, Jafarpour M, Naghshbandi R. The common fimbarie genotyping in Uropathogenic Escherichia coli. Ann Biol Res. 2012;3(10): 4951-4.
17. Momtaz H, Karimian A, Madani M, Dehkordi FS, Ranjbar R, Sarshar M, Souod N. Uropathogenic Escherichia coli in Iran: serogroup distributions, virulence factors and antimicrobial resistance properties. Annals of clinical microbiology and antimicrobials. 2013 Jan; 12(1):8.
18. Martínez-Martínez L, Eliecer Cano M, Manuel Rodríguez-Martínez J, Calvo J, Pascual Á, Hooper, Martens, Wetzstein, Courvalin, Munshi, Rahman. Plasmid-mediated quinolone resistance. Expert review of anti-infective therapy. 2008 Oct 1; 6(5):685-711.
19. Pallecchi L, Bartoloni A, Fiorelli C, Mantella A, Di Maggio T, Gamboa H, Gotuzzo E, Kronvall G, Paradisi F, Rossolini GM. Rapid dissemination and diversity of CTX-M extended-spectrum ß-lactamase genes in commensal Escherichia coli isolates from healthy children from low-resource settings in Latin America. Antimicrobial agents and chemotherapy. 2007 Aug 1; 51(8):2720-5.
20. Marchisio M, Porto A, Joris R, Rico M, Baroni MR, Di Conza J. Susceptibility to ß-lactams and quinolones of Enterobacteriaceae isolated from urinary tract infections in outpatients. Brazilian Journal of Microbiology. 2015 Dec; 46(4):1155-9.
21. Molina-López J, Aparicio-Ozores G, Ribas- Aparicio RM, Gavilanes-Parra S, Chávez-Berrocal ME, Hernández-Castro R, Manjarrez-Hernández HÁ. Drug resistance, serotypes, and phylogenetic groups among uropathogenic Escherichia coli including O25-ST131 in Mexico City. The Journal of Infection in Developing Countries. 2011 Oct 4; 5(12):840-9.
22. Shams F, Hasani A, Pormohammad A, Rezaee MA, Reza M, Nahaie AH, Haghi MH, Gholizade P, Arbatan AE. qnrA implicated quinolone resistance in Escherichia coli and Klebsiella pneumoniae clinical isolates from a University Teaching Hospital. Life Sci J. 2014; 11(12s):1032-5.
23. Bouchakour M, Zerouali K, Claude JD, Amarouch H, El Mdaghri N, Courvalin P, Timinouni M. Plasmid- mediated quinolone resistance in expanded spectrum beta lactamase producing enterobacteriaceae in Morocco. The Journal of Infection in Developing Countries. 2010 Aug 6; 4(12):779-803.
24. Farshad S, Ranijbar R, Japoni A, Hosseini M, Anvarinejad M, Mohammadzadegan R. Microbial susceptibility, virulence factors, and plasmid profiles of uropathogenic Escherichia coli strains isolated from children in Jahrom, Iran. Archives of Iranian Medicine (AIM). 2012 May 1;15(5).