Commentary
A decade ago, Science magazine named cancer immunotherapy as the breakthrough of the year, recognizing its pivotal clinical trials of antibodies designed to inhibit negative regulators of T cell function [1]. Antibody therapies targeting cytotoxic T lymphocyte antigen 4 (CTLA-4) and programmed cell protein 1 (PD-1) initially showed effectiveness against advanced melanoma and have since expanded in use to non-small cell lung, renal cell, and bladder cancers, among others [2-10]. In 2022, lymphocyte activation gene 3 protein (LAG-3) antibody, relatlimab, received FDA approval for unresectable or metastatic melanoma in combination with PD-1 blockade [11]. Despite the proven benefits of these agents, a large number of patients do not respond with positive outcomes. Among patients with previously untreated advanced melanoma, only approximately 30% and 40% achieve long-term disease control with first-line anti-PD-1 monotherapy and anti-PD-1 and anti-CTLA-4 combination therapy, respectively [12-14]. Among the subset of patients with anti-PD-1-refractory disease, only 30% respond to subsequent anti-PD-1 and anti-CTLA-4 combination therapy [15-16]. Furthermore, as the use of immunotherapy in clinical practice matures, the combination regimens that have emerged carry increased risk of toxicity [10]. Achieving the full potential of immunotherapy relies on tailoring treatments to individual patients to maximize effectiveness while minimizing side effects. The success of this endeavor depends on the identification of biomarkers.
Flow cytometry, an important tool in immunology, allows for detailed profiling of the various cell types that make up the immune system. This method generates complex and high-parameter data, characterizing immune cell populations that dynamically shift in response to treatments, and thus requires advanced analytical tools. The TopicFlow computational framework, developed by Peng and colleagues, meets these needs [17]. Utilizing Latent Dirichlet Allocation to analyze flow cytometry data, TopicFlow reveals the dynamics of T cell populations upon treatment exposure and identifies distinct immune response subgroups among patients, thereby deepening our understanding of the immune system's response to immunotherapy. This detailed insight is essential for refining treatment approaches and improving patient outcomes.
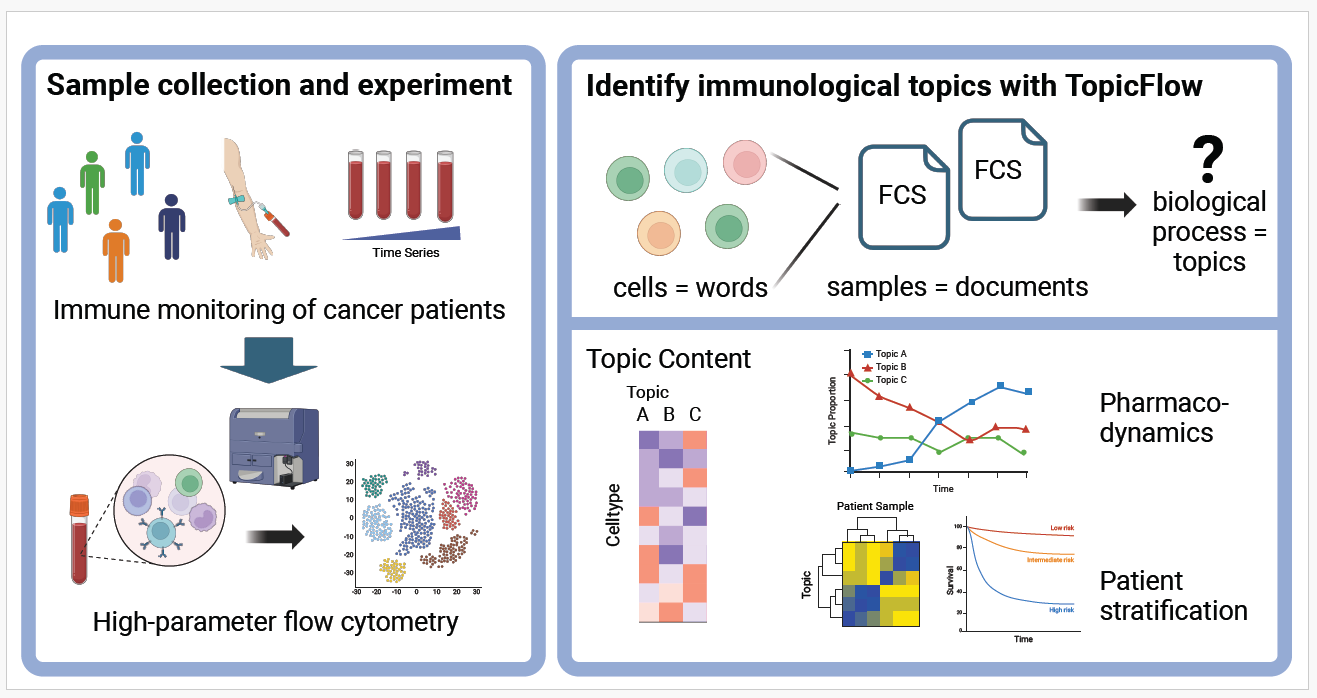
TopicFlow is designed to uncover immunological topics within large-scale flow cytometry data from patients’ samples by conceptualizing cells as words, cell types as terms, and flow samples as documents (Figure 1). Each topic identified through TopicFlow is characterized by a probabilistic distribution across various cell types, delineating the ones that are likely to coexist and contribute to the same biological process. This method, in contrast to traditional approaches that assess cell types individually, allows for a simultaneous evaluation of all cell types and their interactions. By aggregating similar pharmacodynamic trends across multiple cell types, TopicFlow enhances the signal-to-noise ratio. Cell types that co-occur frequently and share similar pharmacodynamic changes tend to be more likely to be grouped into the same topic. In an unsupervised approach, TopicFlow identifies these biologically meaningful topics and provides estimates of the topic proportions for each patient sample. This allows for the stratification of patients into clusters based on the topic compositions. Further exploratory analysis can then be conducted to correlate patient subgroups to clinical outcomes enhancing the understanding of treatment impacts.
Figure 1: TopicFlow identifies biologically interpretable and clinically relevant topics from high-parameter longitudinal flow cytometry data. Created with BioRender.com.
TopicFlow offers several advantages as a computational tool for analyzing flow cytometry data. Primarily, it is an innovative, statistically robust method that can be integrated seamlessly into existing single-cell analysis workflows. The input is a matrix of cell types by sample, derived from cell phenotyping procedures. For users interested in learning more details, we provide a comprehensive tutorial on our GitHub page (https://xiyupeng.github.io/LDA_examples/).
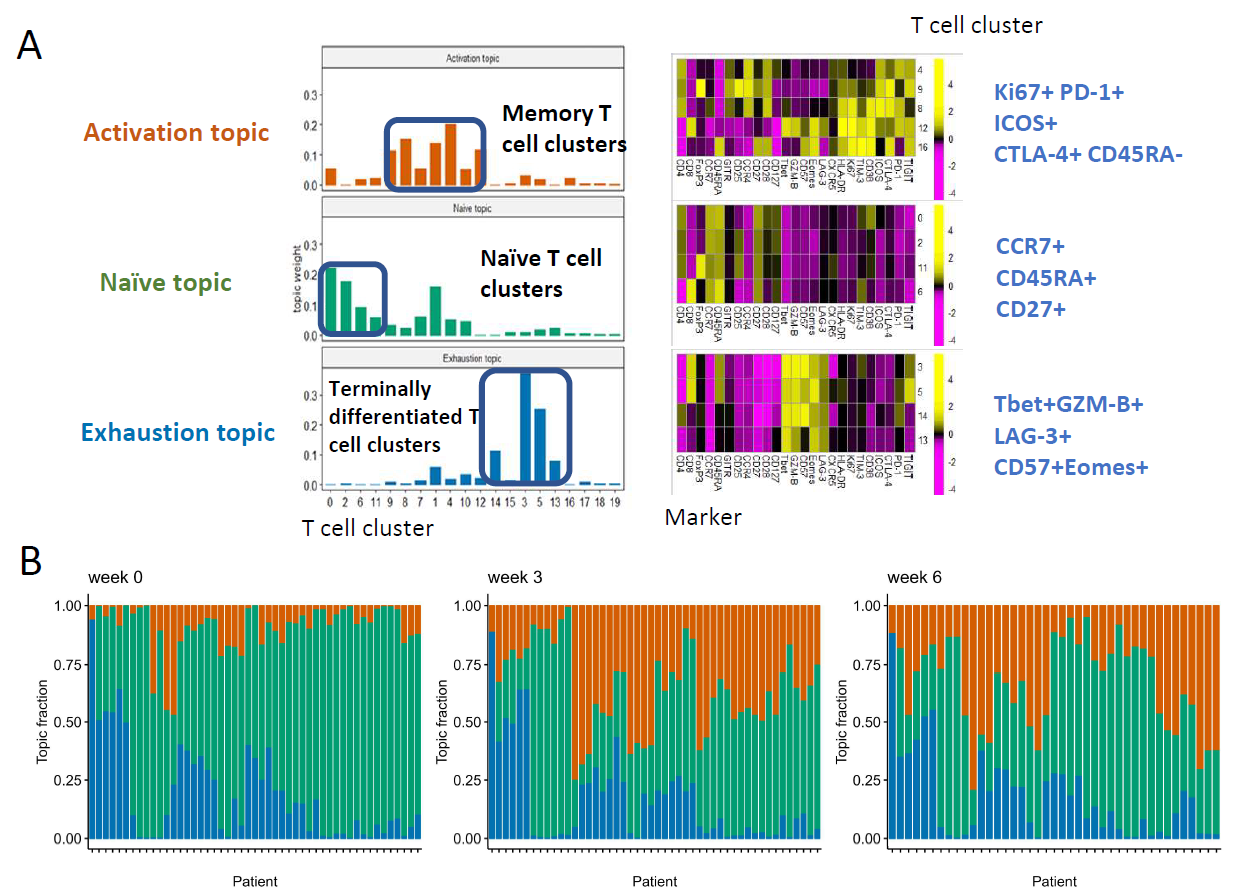
The study by Peng and colleagues illustrates how TopicFlow provides deep insights into highly complex flow cytometry datasets in immuno-oncology research [17]. In their study, they analyzed 138 peripheral blood samples, containing approximately 17 million T cells from 51 patients with melanoma treated with anti-PD-1 and anti-CTLA-4 combination therapy. These samples were collected at three time points: baseline pre-treatment (week 0) and on-treatment (week 3 and week 6). First, T cells were pooled across samples and classified into phenotypes via a graph-based clustering algorithm. TopicFlow was then applied to examine the dynamic changes in these cells, leading to the identification of three distinct T cell topics (Figure 2): (1) an activation topic, predominantly featuring memory T cells (Tcm/Tem), (2) a naive topic, mainly involving naive T cells; and (3) an exhaustion topic, comprised of terminally differentiated T cells (Temra). Each topic corresponds to different T cell phenotypes observed during treatment.
Figure 2: Three topics (activation, naive, exhaustion) were identified by TopicFlow from a longitudinal flow cytometry dataset of melanoma patients treated with anti-PD-1 and anti-CTLA-4 combination therapy [17].
Further analysis of topic proportions across patients and over time recapitulated prior findings regarding treatment resistance and uncovered new insights into the dynamics of immune activation and immune-related toxicity. Earlier studies classified pre-treatment peripheral blood samples from patients into three “immunotypes,” including a “LAG+” immunotype characterized by a high abundance of LAG-3+ CD8+ T cells, which is associated with poorer treatment outcomes [18]. TopicFlow independently recognized a T cell exhaustion topic that aligns closely with the LAG+ immunotype and was present in high proportions in these patients’ samples. The clarity of the topic framework facilitated a deeper examination of the T cell functional characteristics within the LAG+ immunotype, providing opportunity to identify more precise and longitudinally stable biomarkers. The activation topic, which captures the co-expansion of multiple T cell populations including Ki67+ CD8+ T cells, emerged upon treatment, and serves as a complex pharmacodynamic marker of immune response. Relatedly, the naive T cell topic was found to decrease upon treatment, indicating T cell evolution as part of the overall immune response. Interestingly, a higher proportion of the naive topic at baseline was significantly associated with immune-related toxicity during treatment, suggesting its potential as an indicator of adverse immune reactions.
Currently, there are limited computational methods available capable of identifying pharmacodynamic biomarkers from molecular profiles in longitudinally collected patient blood samples. These methods face several challenges and gaps, including the need for integrating temporal data within highly complex single-cell datasets, managing missing data, addressing unevenly spaced time points, and analyzing large-scale data across diverse patient cohorts. Most existing literature has focused on monitoring the pharmacodynamics of limited cell subsets, one at a time, such as the Ki67+ CD8+ T cell subset, longitudinally [19]. Previously Bachireddy et al. [20] utilized a Gaussian process regression model to study temporal dynamics of T cell states in a longitudinal single-cell RNA-seq dataset from a patient cohort undergoing cellular therapy. However, for flow cytometry, TopicFlow appears to be the first computational framework developed for this purpose. It has been used to analyze large-cohort, high-parameter flow cytometry data to identify biomarkers in peripheral blood samples. Moreover, the topic model framework can be extended to other types of single-cell datasets. For instance, we developed SpaTopic [21] for spatial single-cell datasets, integrating spatial information with Latent Dirichlet Allocation to identify tissue architecture from multiplexed imaging.
We intend to validate the promising scientific findings reported by Peng et al. with additional independent datasets. Furthermore, we plan to expand the use of TopicFlow to flow cytometry data from a variety of patient cohorts, encompassing a broader range of clinically relevant scenarios. TopicFlow is not limited to the analysis of flow cytometry data but can also be applied to analyze single-cell RNA-seq data, as demonstrated with a large-scale liver cancer single-cell RNA-seq dataset [22] in the tutorial. The original authors used a highly complex, domain knowledge-intensive method to assign each patient into one of the patient subtypes using ad hoc criteria. In contrast, TopicFlow is more streamlined and structured, requiring no prior knowledge input. It is a more de novo approach for the discovery of cellular subgroups among patients instead of being limited by existing knowledge. A key finding, not discussed in the original study, was the identification of a patient subgroup characterized by a mixture of two cellular topics. Moreover, as an unsupervised technique, TopicFlow does not require a predefined expression profile for each cluster. Instead, it discerns patterns based solely on the pharmacodynamic changes and prevalence of cell subsets across samples. For instance, we highlight that T cell subsets under the same topic not only share similar pharmacodynamic trends, but also show analogous marker expressions [17]. This similarity suggests that these cell subsets are likely involved in similar biological processes during treatment, and grouping these cell types into topics helps clarify the pharmacodynamic profiles associated with specific treatments.
By combining TopicFlow with other computational tools currently under development in our group, we are confident in our capacity to identify blood-based biomarkers that predict outcomes or indicate the pharmacodynamics of various regimens in cancer immunotherapy. Ultimately, these biomarkers may be integrated into immune monitoring and hold potential to inform therapeutic strategies in clinical practice.
Acknowledgements
This work is supported in part by the MSKCC Society, the V Foundation, the Parker Institute for Cancer Immunotherapy, NIH P30 CA008748, NIH R01 CA276286, and the MSK-MIND consortium.
References
2. Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010 Aug 19;363(8):711-23.
3. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012 Jun 28;366(26):2443-54.
4. Brahmer JR, Tykodi SS, Chow LQM, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012 Jun 28;366(26):2455-65.
5. Gettinger SN, Horn L, Gandhi L, Spigel DR, Antonia SJ, Rizvi NA, et al. Overall Survival and Long-Term Safety of Nivolumab (Anti–Programmed Death 1 Antibody, BMS-936558, ONO-4538) in Patients With Previously Treated Advanced Non–Small-Cell Lung Cancer. J Clin Oncol. 2015 Jun 20;33(18):2004-12.
6. Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015 May 21;372(21):2018-28.
7. Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med. 2015 Nov 5;373(19):1803-13.
8. Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet Lond Engl. 2016 May 7;387(10031):1909-20.
9. Motzer RJ, Rini BI, McDermott DF, Redman BG, Kuzel TM, Harrison MR, et al. Nivolumab for Metastatic Renal Cell Carcinoma: Results of a Randomized Phase II Trial. J Clin Oncol Off J Am Soc Clin Oncol. 2015 May 1;33(13):1430-7.
10. Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015 Jul 2;373(1):23-34.
11. Tawbi HA, Schadendorf D, Lipson EJ, Ascierto PA, Matamala L, Castillo Gutiérrez E, et al. Relatlimab and Nivolumab versus Nivolumab in Untreated Advanced Melanoma. N Engl J Med. 2022 Jan 6;386(1):24-34.
12. Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015 Jan 22;372(4):320-30.
13. Robert C, Ribas A, Schachter J, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol. 2019 Sep 1;20(9):1239-51.
14. Wolchok JD, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, et al. Long-Term Outcomes With Nivolumab Plus Ipilimumab or Nivolumab Alone Versus Ipilimumab in Patients With Advanced Melanoma. J Clin Oncol. 2022 Jan 10;40(2):127-137.
15. Olson DJ, Eroglu Z, Brockstein B, Poklepovic AS, Bajaj M, Babu S, et al. Pembrolizumab Plus Ipilimumab Following Anti-PD-1/L1 Failure in Melanoma. J Clin Oncol. 2021 Aug 20;39(24):2647-2655.
16. VanderWalde A, Bellasea SL, Kendra KL, Khushalani NI, Campbell KM, Scumpia PO, et al. Ipilimumab with or without nivolumab in PD-1 or PD-L1 blockade refractory metastatic melanoma: a randomized phase 2 trial. Nat Med. 2023 Sep;29(9):2278-2285.
17. Peng X, Lee J, Adamow M, Maher C, Postow MA, Callahan MK, et al. A topic modeling approach reveals the dynamic T cell composition of peripheral blood during cancer immunotherapy. Cell Rep Methods. 2023;3(8):100546.
18. Shen R, Postow MA, Adamow M, Arora A, Hannum M, Maher C, et al. LAG-3 expression on peripheral blood cells identifies patients with poorer outcomes after immune checkpoint blockade. Sci Transl Med. 2021 Aug 25;13(608):eabf5107.
19. Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017 May;545(7652):60-5.
20. Bachireddy P, Azizi E, Burdziak C, Nguyen VN, Ennis CS, Maurer K, et al. Mapping the evolution of T-cell states during response and resistance to adoptive cellular therapy. Cell Rep. 2021 Nov 9;37(6):109992.
21. Peng X, Smithy JW, Aleynick N, Zhuang M, Li Y, Lee J, et al. 1303 Spatial topic modeling of tumor microenvironment with multiplexed imaging. J Immunother Cancer. 2023 Nov 1;11(Suppl 1):A1448.
22. Xue R, Zhang Q, Cao Q, Kong R, Xiang X, Liu H, et al. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature. 2022 Dec;612(7938):141-7.