Abstract
The expression of small non-coding RNA MicroRNAs (miRNAs) during respiratory viral infections is of critical importance as they are implicated in the viral replication, immune responses and severity of disease pathogenesis. Respiratory viral infections have an extensive impact on human health across the globe. For that is essential to understand the factors that regulate the host response against infections. The differential miRNA pattern induced by respiratory viruses has been reported, including include influenza A virus (IAV), human respiratory syncytial virus (HRSV), human metapneumovirus (HMPV), adenovirus (AdV), and more recently, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection. In this commentary, we highlight the importance of miRNAs identification and the contribution of these molecules in the modulation of the immune response through the upregulation and downregulation of miRNAs expression in different immune and non-immune cells.
Keywords
Respiratory viruses, miRNAs, Respiratory tract, Non-coding RNAs, HMPV, RSV, HRSV
Introduction
MicroRNA (miRNAs) constitute a large family of highly conserved ~21-nucleotide-long small noncoding RNAs. In mammals it is calculated to control ~50% of proteincoding genes by regulation at the post-transcriptional level affecting mRNA degradation [1-3]. This interaction, miRNA-mRNA commonly increases degradation of mRNA by pairing to complementary sequences at the 3´Untranslated Region (3´-UTR) and other important gene sequences repressing translation into protein or promoting mRNA deadenylation [4-6]. Therefore, miRNAs play a role in modulating a variety of biological processes such as development, immune system response and cell death [7-10]. They represent a potential explanation for viral replication and proliferation, host antiviral response, viral pathogenesis in the modulation of inflammatory responses, and their role in the refinement of the innate immune defense against different viruses, diagnostics, and treatment [7,8,11-14].
Differential Induction of miRNAs by HMPV and HRSV in Dendritic Cells
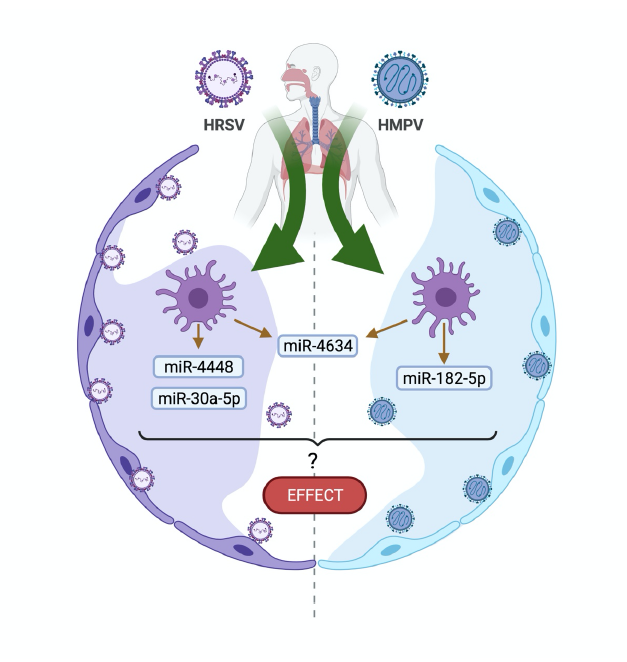
During viral infections some miRNAs are upregulated or downregulated in a large variety of immune cells such as dendritic cells, granulocytes, monocytes, macrophages, and natural killer cells [11,12]. We have previously reported that human metapneumovirus (HMPV) induces a distinct pattern of miRNAs than that of human respiratory syncytial virus (HRSV) in human monocyte-derived dendritic cells (moDCs) [15]. HMPV and HRSV are singlestranded RNA pneumoviruses that cause upper and lower respiratory tract infections (LRTI) in children worldwide [16-21]. We observed that when HMPV infects human moDCs, it induced the expression of hsa-miR-182-5p and hsa-miR-4634 predominantly, while HRSV infection induced a different pattern of miRNAs that included hsamiR- 448, hsa-miR-30a-5p and hsa-miR-4634. Overall, the predominant miRNA induced by both viruses was hsa-miR-4634 (Figure 1) [15]. These findings are relevant since viral infections induce a specific miRNA pattern in different target cells and can differentially modulate the immune response. Future research is warranted to investigate the role of these miRNAs in HMPV and HRSV infection and how the expression of these miRNAs would alter the host immunity as cellular miRNAs induced by other respiratory viruses have been reported to modulate the immune response.
miRNAs and Their Role in Respiratory Viral Infection Immunity
Respiratory viral infections have an extensive impact on human health. Some respiratory viruses have been reported to induce miRNAs implicated in viral replication and immune response modulation[22]. Some examples include influenza A virus (IAV) [7,7,23-28], human respiratory syncytial virus (HRSV) [15,29-34], human metapneumovirus (HMPV) [15,21], adenovirus (ADV) [35], and due to the global health crisis miRNAs have been studied recently in SARS-CoV-2 infection[36].
IAV induces several miRNAs that alter the immune response, including the miR-136 [37], and miR-132- 3p, which targets IRF1, inhibiting IFN response and promoting IAV replication [25]. IAV also induces miR- 29c expression, which correlates with decreased NF- κB activity and expression of TNF-α, IFN-β, IL-6, IL- 1b and IL-8 via the deubiquitinating enzyme A20 [26]. Similarly, IAV H3N2, induces miR-146 a-5p expression and regulates TRAF6 in in human nasal epithelial cells [27]. On the other hand, IAV H5N1 diminishes levels of miR-21-3p in infected patients and A549 cells. miR-21-3p, downregulates IFN response (IFN-β, IFN-α, PKR, MxA, and OAS) by targeting the regulating fibroblast growth factor 2 (FGF2) [28]. It has also been shown that IAV H5N1 downregulates the miRNA family (30-a, -b, -c, -d, and -e) in cell lines. The expression of SOCS1 and SOCS2 is downregulated by IAV H5N1, abrogating the IFN/JACK/ STAT pathway’s inhibition effect. miRNA 30 family also inhibits the expression of the E3 ubiquitin ligase NEDD4, which negatively regulates IFITM3 [38].
HRSV infection is the main cause of hospitalization due to respiratory disease in infants and young children [31,32,39]. It is one of the respiratory viruses most extensively studied for its induction of miRNAs. Data from blood samples identified 168 miRNAs in patients with severe HRSV-associated pediatric pneumonia and 131 miRNAs in patients with mild HRSV-associated pediatric pneumonia, which might be involved in the NF-κB and MAPK signaling pathways, both essential components of the immune response [29]. When blood samples from children with severe HRSV-caused pneumonia were analyzed, four miRNAs (miR-1271-5p, miR-10a-3p, miR- 125b-5p, and miR-30b-3p) were found upregulated [29]. However, when blood samples from patients hospitalized with HRSV infection were analyzed, a different pattern of overexpressed miRNA was observed (miR-106b-5p, miR- 20b-5p, and miR-342-3p) [33], suggesting that the host response shapes the induction of the miRNA pattern.
The expression of miRNAs induced by HRSV has also been studied in primary cells and cell lines. Our reported study focused mainly on peripheral blood monocytederived dendritic cells (MoDC), where we observed HRSV infection upregulates the expression of miR-4448, miR-30a-5p, and miR-4634 [15]. Thornburg et al. has also reported the expression of miR-Let-7b by HRSV in moDCs [31]. Work on A549 cells, a human lung-derived epithelial cell line, has shown the upregulation of miR-let- 7f, miR-24, miR-337-3p, miR-26b, and miR-520a-5p [32]. Additionally, in NHBE cells (primary human bronchial/ tracheal epithelial cells), HRSV increased levels of miRlet- 7c, miR-let-7i, and miR-30b [31]. These observations indicate that the miRNA pattern induced by the virus depends on the type of cell infected.
Downregulation of miRNAs due to HRSV infection also has been reported. In A549 cells, HRSV repressed miR-198 and miR-595 [32]; while in blood samples from patients with HRSV infection, a different pattern of miRNAs was downregulated (miR-320e, miR-320d, miR-877-5p, miR-122-5p, and miR-92b-5p) [33]. In nasal samples from infected patients, however, miR-140-5p was found reduced [34].
Although the target genes affected by the HRSV-induced miRNAs and the mechanisms of the alterations in the immune response are not fully understood, some partial mechanisms and target genes have been identified. miR- 30b appears to be induced through the NF-κB pathway [40], while miR-Let-7b appears to be increased by IFN-β [31]. In A549 cells, miR-let-7f was identified as a regulator of CCL7 and SOCS3, genes involved in cytokine response against viruses, in a mechanism involving the HRSV attachment protein [32]. The upregulated miRNAs (miR-1271-5p, miR-10a-3p, miR-125b-5p, and miR-30b- 3p) found in blood samples from infected patients have predicted target genes involved in the NF-kB and the MAPK signaling pathway, and T cell receptor signaling proteins (TNFRSF19, HMOX1, TLR4, LCK, and ZAP70) [29]. Likewise, overexpression of miR-106b-5p, miR- 20b-5p, miR-342-3p, miR-320e, miR-320d, miR -877- 5p, miR-122-5p, and miR-92b-5p may be involved in inflammatory and immune responses by likely targeting the activation pathways of TGF-β, insulin, and T and B cell receptors [33]. On the other hand, the reduced expression of miR-140-5p in nasal and blood samples of patients with HRSV infection led to the downregulation of TNF-α, and IL-1β, -6 and -8, mediated by IFN-α (in BEAS-2B cells). In bronchial epithelial cells, the downregulation of miR-34b/ c-5p resulted in increased expression of Muc5ac, probably through of c-Jun activation pathway [39].
Human metapneumovirus (HMPV) belongs to the Pneumoviridae family and is the first human member of the Metapneumovirus genus [18,20]. This virus presents similar pathology and clinical symptoms as the ones of HRSV. In both cases coughing, bronchitis, bronchiolitis and respiratory manifestations related to the absence of airflow are seen [41,42]. HMPV affects all age groups worldwide, with a risk of sever disease found in immunocompetent patients, children and elderly [43]. HMPV infection alters the expression of multiple cellular miRNAs in several cells [21]. We reported that HMPV upregulated significantly the expression of miR-182-5p, and miR-4634 in moDCs, where we highlighted the upregulation of miR-182-5p [15], which might contribute to the imbalance of Th17 cells [44] as shown in unrelated models where miR-182- 5p triggers the secretion of cytokines related to a Th2-like profile, leading to high recruitment of neutrophils in the lung and secretion of cytokines such as IL-13 and IL-5 [16], and protects against different intracellular pathogens, increasing the expression of interferons and other proinflammatory cytokines [45,46]. In A549 cells, the miRNAs expression pattern is different. HMPV induced the expression of miR-Let-7f, miR-192, miR-374a, and miR-452, which are upregulated by HMPV, where miRLet- 7f appears to favors the replication of HMPV [21] through an unknown mechanism.
Like HMPV and HRSV, another cause of pneumonia in infants and children is the human adenovirus (HAdV) infection. The expression profile of whole blood samples from healthy and infected patients with HAdV, indicated the expression of 3 upregulated miRNAs (miR-127-3p, miR-493-5p, and miR-409-3p). Such miRNAs were found to target PSMB5, ITGA6, MYCBP2, TCF7L2, UBE2V2, HIPK1, UBE2D2, and KANSL1genes, mostly involved in cellular processes, and MAPK and Ras signaling [35].
In search of therapeutic targets against the severe acute respiratory syndrome coronavirus-2 (SARSCoV- 2) infection, miRNAs induced by this virus have also been investigated [47]. Lu et al. studied the miRNAs differentially expressed in blood samples from patients with COVID-19, and they found that miR-16-2-3p, miR- 6501-5p, and miR-618, miR183-5p, miR-627-5p, and miR- 144-3p were upregulated in COVID-19 positive patients compared to healthy individuals [47]. Analyses in silico on the SARS-CoV-2 genome sequence have revealed matching of viral genes with some elements involved in the immune response. Specifically, the protein S exhibits miRNA-like sequences that match with IFNA7 and IRF1; but, if these interactions occur in the infection, and their biological implications remained to be explored [48]. Likewise, Aydemir et al. found 40 SARS-CoV-2 miRNAs, which target genes involved in the immune response through NF-κB, JAK/STAT and TGF-β signaling pathways. However, the in vivo relevance of these molecules is so far unknown [49].
Conclusions and Perspectives
The upregulation or downregulation of miRNAs during respiratory viral infections can act differently in the host by altering multiple processes, including the immune response. The presence of specific miRNAs can modulate the immune response by inhibiting or activating genes involved in the expression of inflammatory cytokines, IFN response, cellular receptors, and other immune processes. On the other hand, miRNA expression can affect viral replication by increasing or decreasing the viral gene expression. Strikingly, miRNAs can have a paracrine effect on other non-infected cells to activate the immune response. Future research is warranted to investigate the profiles of miRNA expression induced by viral infections in the respiratory tract in the different immune and nonimmune cells to understand more on how miRNAs are involved in viral pathogenesis. Furthermore, it is observed that viruses can induce different miRNA profiles but exert a similar effect on the host immune response. That phenomenon may result from having different miRNAs targeting the expression of distinct proteins, but regulating similar immune pathways, e.g., different miRNAs targeting diverse cellular receptors that trigger an IFN response. That scenario could represent a viral survival strategy allowing the virus to replicate more efficiently. Therefore, further research on the functional validation of the molecules targeted by the diverse miRNAs is essential to fully understand the regulatory mechanisms induced by viral infections.
Although the use of miRNAs as therapeutic agents remains largely under development, there are promising data using specific miRNA antagonists (antagomirs) to treat viral infections. That is indicated by the clinical trials testing the oligonucleotide Miravirsen (SPC3649) targeting miR-122 for the treatment of hepatitis C infection [50,51]. However, more information on the identification of miRNAs and their targets, as well as on the characterization of the underlying molecular mechanisms of miRNA-mRNA regulation during viral infections is critical to design improved antiviral therapies for respiratory viral infections.
| VIRUS | miRNA | EFFECT | CELL | REFS. |
|---|---|---|---|---|
| IAV H1N1 | ↑miR-132-3p | Suppresses type I IFN response and promotes IAV replication |
A549 cells | [25] |
| ↑miR- 29c | Decreases NFκB, TNF-α, IFN-β, IL-6, IL-1β and IL-8 | [26] | ||
| IAV H3N2 | ↑mir-146 a-5p | Regulates TRAF6 | Human nasal epithelial cells | [27] |
| IAV H5N1 | ↑miR-136 | Acts as an immune agonist of RIG-I, thus leads to IL-6 and IFN-β accumulation |
A549 cells | [37] |
| ↑miR-21-3p | Decreases the levels of IFN-β, IFN-α, PKR, MxA, and OAS | [28] | ||
| ↓miR-30a,-b,-c,- d,-e. | Modulates SOCS1, SOCS3, and NEDD4 to impair the host antiviral response |
[38] | ||
| HRSV | ↑miR-1271-5p | Undefined | Whole blood samples | [29] |
| ↑miR-10a-3p | ||||
| ↑miR-125b-5p | ||||
| ↑miR-30b-3p | ||||
| ↑miR-106b-5p | Undefined | Peripheral blood | [33] | |
| ↑miR-20b-5p | ||||
| ↑miR-342-3p | ||||
| ↓miR-320e | ||||
| ↓miR -320d | ||||
| ↓miR -877-5p | ||||
| ↓miR-122-5p | ||||
| ↓miR-92b-5p | ||||
| ↓miR-34b/c-5p | Induces Muc5ac expression | Human bronchial epithelial cells | [39] | |
| ↑miR-Let-7b | Regulates IFN-β response | MoDCs | [31] | |
| ↑miR-4448 | Undefined | MoDCs | [15] | |
| ↑miR-30a-5p | ||||
| ↑miR-4634 | ||||
| ↑miR-let-7f | Undefined | A549 cells | [32] | |
| ↑miR-24 | ||||
| ↑miR-337-3p | ||||
| ↑miR-26b | ||||
| ↑miR-520a-5p | ||||
| ↑miR-let-7c | Undefined | Normal human bronchial epithelial cells | [31] | |
| ↑miR-let-7i | Regulates IFN-β response | |||
| ↑miR-30b | Undefined | |||
| ↓miR-198 | Undefined | A549 cells | [32] | |
| ↓miR-595 | ||||
| HMPV | ↑miR-182-5p | Undefined | MoDCs | [15] |
| ↑miR-4634 | ||||
| ↑miR-Let-7f | Regulates viral replication | A549 cells | [21] | |
| ↓miR-192 | Undefined | |||
| ↓miR-374a | ||||
| ↑miR-452 | ||||
| HAdV | ↑miR-127-3p | Undefined | Whole blood samples | [35] |
| ↑miR-493-5p | ||||
| ↑miR-409-3p | ||||
| SARS-CoV-2 | ↑miR-16-2-3p | Undefined | Whole blood samples | [47] |
| ↑miR-6501-5p | ||||
| ↑miR-618 | ||||
| ↓miR183-5p | ||||
| ↓miR-627-5p | ||||
| ↓miR144-3p |
Funding
This research was funded by a grant from National Institute of General Medical Sciences (P20GM130555) and R03 AI081171 from the U.S. National Institutes of Health and the Louisiana State University School of Veterinary Medicine Competitive Research Program (to A.G.-P.).
Conflicts of Interest
The authors declare no conflict of interest.
References
2. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics. 2010 Sep;11(9):597-610.
3. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan 23;116(2):281-97.
4. Cullen BR. Viral and cellular messenger RNA targets of viral microRNAs. Nature. 2009 Jan;457(7228):421-5.
5. Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proceedings of the National Academy of Sciences. 2010 Apr 6;107(14):6328-33.
6. Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Research. 2018 Jan 4;46(D1):D296-302.
7. Brogaard L, Larsen LE, Heegaard PM, Anthon C, Gorodkin J, Dürrwald R, et al. IFN-λ and microRNAs are important modulators of the pulmonary innate immune response against influenza A (H1N2) infection in pigs. PLoS One. 2018 Apr 20;13(4):e0194765.
8. Jafarinejad-Farsangi S, Jazi MM, Rostamzadeh F, Hadizadeh M. High affinity of host human microRNAs to SARS-CoV-2 genome: An in silico analysis. Non-coding RNA Research. 2020 Dec 1;5(4):222-31.
9. Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Letters. 2005 Oct 31;579(26):5911-22.
10. Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005 Nov 1;132(21):4653-62.
11. Hou J, Wang P, Lin L, Liu X, Ma F, An H, et al. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. The Journal of Immunology. 2009 Aug 1;183(3):2150-8.
12. Li Z, Tzeng CM. Integrated analysis of miRNA and mRNA expression profiles to identify miRNA targets. In: mRNA Decay. New York: Humana Press. 2018; pp. 141- 148.
13. Trobaugh DW, Klimstra WB. MicroRNA regulation of RNA virus replication and pathogenesis. Trends in Molecular Medicine. 2017 Jan 1;23(1):80-93.
14. Manjarrez-Zavala ME, Rosete-Olvera DP, Gutiérrez- González LH, Ocadiz-Delgado R, Cabello-Gutiérrez C. Pathogenesis of viral respiratory infection. Respiratory Disease and Infection: A New Insight. 2013 Feb 6;1.
15. Baños-Lara MD, Zabaleta J, Garai J, Baddoo M, Guerrero-Plata A. Comparative analysis of miRNA profile in human dendritic cells infected with respiratory syncytial virus and human metapneumovirus. BMC Research Notes. 2018 Dec;11(1):1-7.
16. Lay MK, Céspedes PF, Palavecino CE, León MA, Díaz RA, Salazar FJ, et al. Human metapneumovirus infection activates the TSLP pathway that drives excessive pulmonary inflammation and viral replication in mice. European journal of immunology. 2015 Jun;45(6):1680- 95.
17. Van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nature medicine. 2001 Jun;7(6):719-24.
18. van den Hoogen BG, Bestebroer TM, Osterhaus AD, Fouchier RA. Analysis of the genomic sequence of a human metapneumovirus. Virology. 2002 Mar 30;295(1):119-32.
19. Goutagny N, Jiang Z, Tian J, Parroche P, Schickli J, Monks BG, et al. Cell type-specific recognition of human metapneumoviruses (HMPVs) by retinoic acid-inducible gene I (RIG-I) and TLR7 and viral interference of RIG-I ligand recognition by HMPV-B1 phosphoprotein. The Journal of Immunology. 2010 Feb 1;184(3):1168-79.
20. Haas LE, Thijsen SF, Van Elden L, Heemstra KA. Human metapneumovirus in adults. Viruses. 2013 Jan;5(1):87-110.
21. Deng J, Ptashkin RN, Wang Q, Liu G, Zhang G, Lee I, et al. Human metapneumovirus infection induces significant changes in small noncoding RNA expression in airway epithelial cells. Molecular Therapy-Nucleic Acids. 2014 Jan 1;3:e163.
22. Leon-Icaza SA, Zeng M, Rosas-Taraco AG. microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA. 2019 Dec;1(1):1-7.
23. Ingle H, Kumar S, Raut AA, Mishra A, Kulkarni DD, Kameyama T, et al. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Science Signaling. 2015 Dec 8;8(406):ra126.
24. Skovgaard K, Cirera S, Vasby D, Podolska A, Breum SØ, Dürrwald R, et al. Expression of innate immune genes, proteins and microRNAs in lung tissue of pigs infected experimentally with influenza virus (H1N2). Innate Immunity. 2013 Oct;19(5):531-44.
25. Zhang F, Lin X, Yang X, Lu G, Zhang Q, Zhang C. MicroRNA-132-3p suppresses type I IFN response through targeting IRF1 to facilitate H1N1 influenza A virus infection. Bioscience Reports. 2019 Dec 20;39(12).
26. Zhang X, Dong C, Sun X, Li Z, Zhang M, Guan Z, et al. Induction of the cellular miR-29c by influenza virus inhibits the innate immune response through protection of A20 mRNA. Biochemical and Biophysical Research Communications. 2014 Jul 18;450(1):755-61.
27. Deng Y, Yan Y, Tan KS, Liu J, Chow VT, Tao ZZ, et al. MicroRNA-146a induction during influenza H3N2 virus infection targets and regulates TRAF6 levels in human nasal epithelial cells (hNECs). Experimental Cell Research. 2017 Mar 15;352(2):184-92.
28. Shi J, Feng P, Gu T. MicroRNA-21-3p modulates FGF2 to facilitate influenza A virus H5N1 replication by refraining type I interferon response. Bioscience Reports. 2020 May 29;40(5).
29. Zhang X, Huang F, Yang D, Peng T, Lu G. Identification of miRNA-mRNA Crosstalk in Respiratory Syncytial Virus-(RSV-) Associated Pediatric Pneumonia through Integrated miRNAome and Transcriptome Analysis. Mediators of Inflammation. 2020 May 1;2020.
30. Groskreutz DJ, Monick MM, Yarovinsky TO, Powers LS, Quelle DE, Varga SM, et al. Respiratory syncytial virus decreases p53 protein to prolong survival of airway epithelial cells. The Journal of Immunology. 2007 Sep 1;179(5):2741-7.
31. Thornburg NJ, Hayward SL, Crowe Jr JE. Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF-κB. MBio. 2012 Dec 18;3(6):e00220-12.
32. Bakre A, Mitchell P, Coleman JK, Jones LP, Saavedra G, Teng M, et al. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. The Journal of General Virology. 2012 Nov;93(11):2346.
33. Wang S, Liu P, Yang P, Zheng J, Zhao D. Peripheral blood microRNAs expression is associated with infant respiratory syncytial virus infection. Oncotarget. 2017 Nov 14;8(57):96627.
34. Zhang Y, Shao L. Decreased microRNA?140?5p contributes to respiratory syncytial virus disease through targeting Toll?like receptor 4. Experimental and Therapeutic Medicine. 2018 Aug 1;16(2):993-9.
35. Huang F, Zhang J, Yang D, Zhang Y, Huang J, Yuan Y, et al. MicroRNA expression profile of whole blood is altered in adenovirus-infected pneumonia children. Mediators of inflammation. 2018 Oct 14;2018 :2320640.
36. Mirzaei R, Mahdavi F, Badrzadeh F, Hosseini-Fard SR, Heidary M, Jeda AS, et al. The emerging role of microRNAs in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. International Immunopharmacology. 2021 Jan;90:107204.
37. Zhao L, Zhu J, Zhou H, Zhao Z, Zou Z, Liu X, et al. Identification of cellular microRNA-136 as a dual regulator of RIG-I-mediated innate immunity that antagonizes H5N1 IAV replication in A549 cells. Scientific Reports. 2015 Oct 9;5(1):1-3.
38. Lin X, Yu S, Ren P, Sun X, Jin M. Human microRNA-30 inhibits influenza virus infection by suppressing the expression of SOCS1, SOCS3, and NEDD4. Cellular Microbiology. 2020 May;22(5):e13150.
39. Du X, Yang Y, Xiao G, Yang M, Yuan L, Qin L, et al. Respiratory syncytial virus infection-induced mucus secretion by down-regulation of miR-34b/c-5p expression in airway epithelial cells. Journal of Cellular and Molecular Medicine. 2020 Nov;24(21):12694-705.
40. Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009; 1(6): a001651.
41. Schildgen V, van den Hoogen B, Fouchier R, Tripp RA, Alvarez R, Manoha C, et al. Human Metapneumovirus: lessons learned over the first decade. Clinical Microbiology Reviews. 2011 Oct;24(4):734-54.
42. Huck B, Neumann-Haefelin D, Schmitt-Graeff A, Weckmann M, Mattes J, Ehl S, et al. Human metapneumovirus induces more severe disease and stronger innate immune response in BALB/c mice as compared with respiratory syncytial virus. Respiratory Research. 2007 Dec;8(1):1.
43. Cifuentes-Muñoz N, Branttie J, Slaughter KB, Dutch RE. Human metapneumovirus induces formation of inclusion bodies for efficient genome replication and transcription. Journal of Virology. 2017 Nov 30;91(24):e01282-17.
44. Hua M, Li J, Wang C, Shao L, Hou M, Peng J, et al. Aberrant expression of microRNA in CD4+ cells contributes to Th17/Treg imbalance in primary immune thrombocytopenia. Thrombosis Research. 2019 May 1;177:70-8.
45. Gregory DJ, Kramnik I, Kobzik L. Protection of macrophages from intracellular pathogens by miR-182-5p mimic—a gene expression meta-analysis approach. The FEBS Journal. 2018 Jan 1;285(2):244-60.
46. Liao S, Bao X, Liu T, Lai S, Li K, Garofalo RP, Casola A. Role of Retinoic Acid Inducible Gene-I in Human Metapneumovirus-induced Cellular Signaling. The Journal of General Virology. 2008 Aug;89(8):1978.
47. Li C, Hu X, Li L, Li JH. Differential microRNA expression in the peripheral blood from human patients with COVID-19. Journal of Clinical Laboratory Analysis. 2020 Oct;34(10):e23590.
48. Demongeot J, Seligmann H. SARS-CoV-2 and miRNA-like inhibition power. Medical Hypotheses. 2020 Nov 1;144:110245.
49. Aydemir MN, Aydemir HB, Korkmaz EM, Budak M, Cekin N, Pinarbasi E. Computationally predicted SARSCOV- 2 encoded microRNAs target NFKB, JAK/STAT and TGFB signaling pathways. Gene Reports. 2021 Mar 1;22:101012.
50. Bonneau E, Neveu B, Kostantin E, Tsongalis GJ, De Guire V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC. 2019 Jun;30(2):114.
51. Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. New England Journal of Medicine. 2013 May 2;368(18):1685-94.