Abstract
Accumulating evidence from recent research offers new perspectives on the functions of long non-coding RNAs (lncRNAs) in immunooncology. In addition to modulating the aggressiveness of cancer cells, lncRNAs are essential players in regulating various immune cells and stromal cells, playing a role in reshaping the tumor microenvironment and affecting anti-tumor immunity. The insightful discoveries on the role of lncRNAs in immuno-oncological activities indicate the prognostic value of lncRNA markers. Here, we present an overview of the roles of lncRNAs derived from different cell types in the tumor microenvironment, that is, immune cells, tumor cells, and stromal cells, and summarize their functional characterization and mechanisms in immuno-oncological activities. We also discuss the opportunities and challenges of single-cell-based technologies for analyzing the cellular function of immune-related lncRNAs.
Keywords
Long noncoding RNA, Cancer, Immune cells, Tumor microenvironment, Immuno-oncology, Immune escape, Cancer-immune cycle, lncRNA signature
Introduction
Long non-coding RNA (lncRNAs) are RNA transcripts that are longer than 200 nucleotides [1]. Initially, lncRNAs were considered as “waste” resulting from messenger RNA transcription due to their inability to encode proteins [2]. However, in recent decades, it has been discovered that some lncRNAs have peptide-encoding ability [1], and extensive research has revolutionized our understanding of the function of lncRNAs, especially in cancers [3-5]. In particular, recent advances in the research on lncRNAs have uncovered their substantial role in shaping the tumor microenvironment (TME) in various malignancies, the development and progression of which are controlled by the dynamic interactions between cancer cells and the TME.
In the TME, a complex mixture of non-malignant components, such as immune cells, fibroblasts, and extracellular matrix, is extensively involved in cancer immunity. The cancer-immunity cycle consists of a series of complex steps. Initially, antigens are released, and these are recognized by dendritic cells (DCs) and presented on major histocompatibility class I (MHCI) molecules. Next, the antigen-MHCI complex is specifically recognized by CD8+ T cells, and this triggers CD8+ T cell activation. On activation, CD8+ T cells migrate into the tumor, where they recognize and kill cancerous cells [6]. Other cells in the TME also play pivotal roles in regulating anti-tumor immunity, including immune cells such as regulatory T cells (Treg cells), macrophages, and myeloid-derived suppressor cells (MDSCs), as well as stromal cells such as fibroblasts.
LncRNAs have been found to regulate the behavior of cancer cells, such as invasion, proliferation, and epithelial-mesenchymal transition (EMT), and they also serve as key regulators in immune cells, such as T cells, B cells, macrophages, and dendritic cells, which are closely associated with anti-tumor activities and immune evasion (as discussed above). In this review, we discuss how immune-related lncRNAs derived from different cell types in the TME play promoting or suppressive roles in cancer development, describe the regulatory functions and mechanisms of these lncRNAs, and highlight their potential clinical value. The immune-related lncRNAs discussed here not only include those expressed by immune cells and involved in regulating immune cell activities, but also include those which participate broadly in the regulation of cancer immunity. The latter lncRNAs can be expressed by either immune cells or non-immune cells, including cancer cells and stromal cells.
Multicellular Functions of lncRNAs in the TME
LncRNAs have been implicated in a wide range of functions in the TME. It has been widely demonstrated that some lncRNAs exert tumor promotion or suppression functions not only in cancer cells, but also in non-malignant cells. For instance, lncRNAs, such as PCAT6, UCA1, NKILA, NEAT1, LUCAT1, HOTAIR, and NRON, which are involved in oncogenic or anti-tumor activities in cancer cells, have also been found to play important roles in various types of immune cells (Table 1). The roles of these lncRNAs are discussed in more detail below.
|
LncRNA |
Cell Type |
Disease |
Function |
Mechanism |
References |
|
PCAT6 |
Cancer cell |
NSCLS |
Promotes cell growth |
represses LATS2 via the epigenetic repressor EZH2 |
[7] |
|
Cancer cell |
Breast cancer |
Promotes cell proliferation, migration and angiogenesis |
upregulated by VEGF secreted by M2 macrophage and induces the expression of VEGFR2 via ceRNA and deubiquitylation mechanism |
[9] |
|
|
Cancer cell |
Prostate cancer |
Promotes cell proliferation, migration and invasion |
METTL3-mediated m6A modification |
[8] |
|
|
Macrophage |
Cholangiocarcinoma |
Promotes M2 polarization of macrophage |
miR-326 and RhoA-ROCK pathway |
[10] |
|
|
SNHG1 |
Cancer cell |
Gastric Cancer |
Promotes cell proliferation |
promotes DNMT1 expression |
[11] |
|
CD4+ T cell |
Breast cancer |
Regulates Tregs differentiation |
miR-448/IDO |
[12] |
|
|
NKILA |
Cancer cell |
Breast cancer |
Inhibits EMT |
Inhibits NF-κB activity |
[17] |
|
Cancer cell |
Breast cancer |
Suppresses metastasis |
Prevents over-activation of NF-κB pathway |
[16] |
|
|
T cell |
Breast cancer |
Induces apoptosis of T cells and inhibits CTL infiltration |
Inhibits NF-κB activity |
[18] |
|
|
NEAT1 |
Cancer cell |
NSCLC |
Promotes cell proliferation |
miR-377-3p/E2F3 |
[13] |
|
Cancer cell |
Prostate cancer |
Promotes cancer progression and induces resistance to androgen or AR antagonists |
increased H3K4Me3 and H3AcK9 deposits on the PSMA promoter and transcriptionally activated PSMA |
[14] |
|
|
CD8+ T cell |
Hepatocellular carcinoma |
Inhibits CD8+ T cell apoptosis and enhances cytolytic activity |
miR-155 |
[15] |
|
|
Myeloid cell |
Acute promyelocytic leukemia |
Inhibits myeloid differentiation |
/ |
[126] |
|
|
DC |
Autoimmune disease |
Induces immune tolerance in DC |
miR-3076-3p/NLRP3 |
[127] |
|
|
LUCAT1 |
Cancer cell |
Colorectal cancer |
Promotes cell proliferation and chemotherapy resistance |
Interacts with PTBP1 to alter alternative splicing of DNA damage related genes |
[128] |
|
Cancer cell |
Breast cancer |
Promotes breast cancer stemness |
miR-5582-3p/TCF7L2 |
[129] |
|
|
Myeloid cell |
/ |
Inhibits immune response |
Interacts with STAT1 to inhibit ISGs |
[130] |
|
|
HOTAIR |
Cancer cell |
Laryngeal Carcinoma |
Promotes cell proliferation and PD-L1 expression |
miR-30a-5p/GRP78/PD-L1 |
[131] |
|
Cancer cell |
Glioma |
Upregulates PD-L1 and inhibits T cells activation |
NF-κB pathway |
[132] |
|
|
Macrophage |
/ |
Promotes the NF-κB-mediated inflammatory pathway |
Activates NF-κB and upregulates IL-6 and iNOS expression via facilitating the degradation of IκBα. |
[133] |
|
|
Macrophage |
/ |
Regulates glucose metabolism |
NF-κB pathway |
[134] |
|
|
NRON |
Cancer cell |
Hepatocellular carcinoma |
Suppresses cell growth and metastasis |
Inhibits EMT |
[135] |
|
T cell |
/ |
Represses nuclear factor of activated T cells |
/ |
[136] |
The lncRNA PCAT6 has been identified as an oncogene in various types of cancers. For example, in non-small-cell lung cancer (NSCLC) and prostate cancer, PCAT6 induces an increase in the proliferation and metastasis of cancer cells via epigenetic modifications [7, 8]. In triple-negative breast cancer, PCAT6 is stimulated by vascular epithelial growth factor (VEGF) secreted by M2 macrophages and induces VEGFR2 expression via a competing endogenous RNA (ceRNA) and deubiquitylation mechanism, thus promoting angiogenesis [9]. Subsequent research has shown that PCAT6 plays a bigger role in immune cells. That is, overexpression of PCAT6 contributes to the accumulation of reactive oxygen species via the miR-326 and RhoA-ROCK pathway and leads to mitochondrial and metabolic function disorders, thus promoting macrophage M2 polarization, which is critical for immune suppression [10] (Figure 1). The lncRNA small nucleolar RNA host gene 1 (SNHG1) also acts as an oncogene in cancer cells [11], while in Treg cells, SNHG1 promotes cell differentiation by sponging miR-448 and upregulating indoleamine 2,3-dioxygenase (IDO) [12]. Further, the lncRNA nuclear enriched abundant transcript 1 (NEAT1) accelerates tumor growth in NSCLC and prostate cancers [13,14], as well as acts as a tumor promotor by accelerating CD8+ T-cell apoptosis and inhibiting cytolytic activity [15].
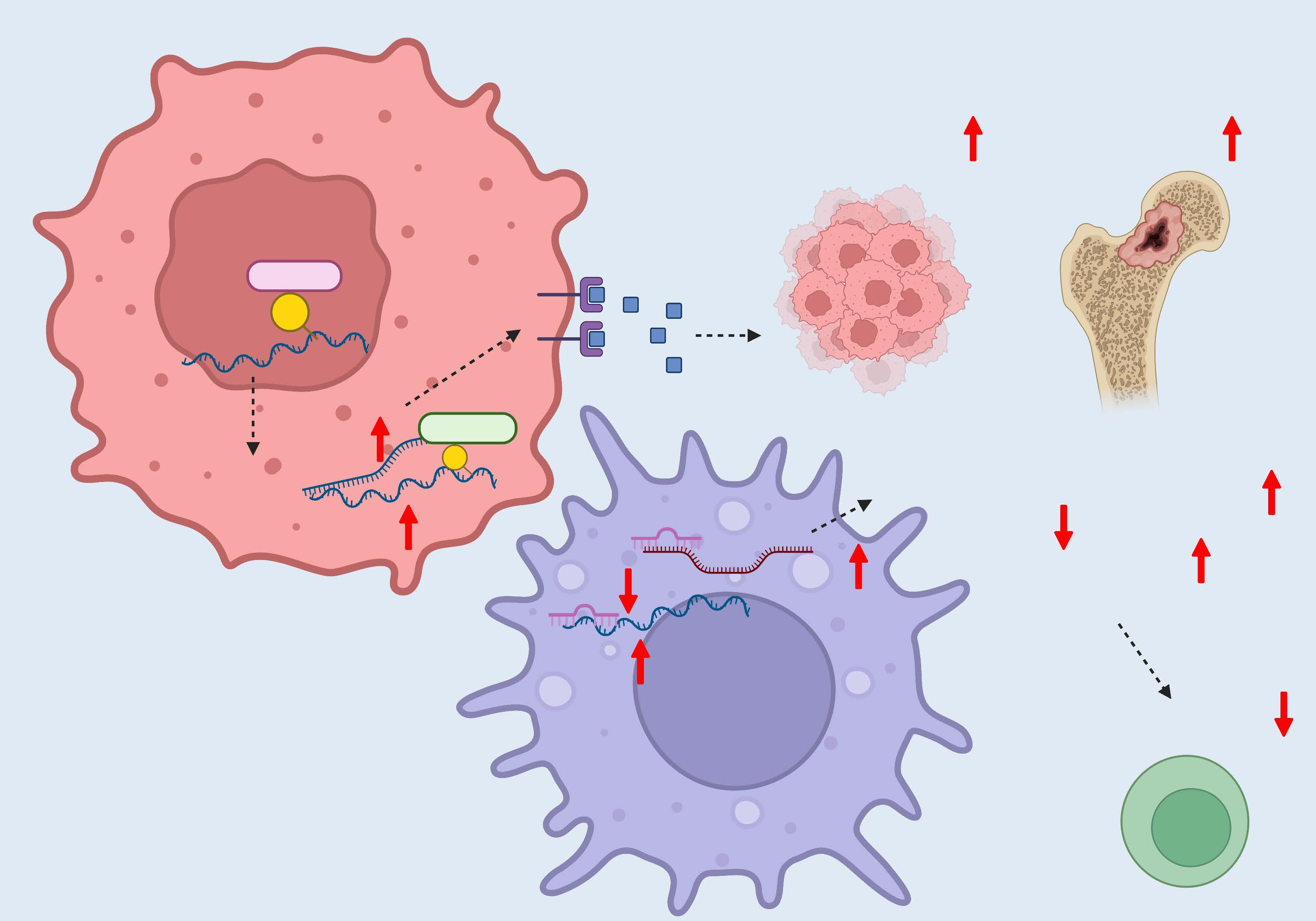
Figure 1. Functions and mechanisms of lncRNA PCAT6 in the TME.
In some cases, lncRNAs play opposing roles as cancer-promoting and cancer-suppressing molecules in malignant and non-malignant cells, respectively. For example, in breast cancer, NF-κB-interacting lncRNA (NKILA), which is upregulated by NF-κB and forms a stable complex by interacting with NF-κB/IκB, plays anti-tumor roles by preventing hyperactivation of the NF-κB pathway in tumor cells. The degradation of NKILA by miR-103/107 targeting activates NF-κB signaling and increases invasiveness in breast cancer cell lines [16]. Similarly, it has been reported that TGF-β-induced NKILA suppresses EMT in breast cancer by negative regulation of the TGF-β-induced NF-κB pathway [17]. On the other hand, the lncRNA NKILA also participates in immune escape in the TME. NKILA expression is upregulated in cytotoxic T lymphocytes (CTLs) and Th1 cells, which suppresses NF-κB activity and sensitizes T cells [24]. These changes control immune escape by inducing apoptosis of T cells and inhibiting CTL infiltration in the cancer [18].
Collectively, the findings so far indicate that lncRNAs may play different roles in the TME depending on cell type and shed light on the multifaceted functions of lncRNAs and the complex regulation of lncRNAs. Thus, not only lncRNAs derived from malignant cells, but also those expressed in immune or stromal cells, could function as tumor suppressors or promotors, and mediate immune activities in the TME.
Tumor Cell-derived lncRNAs
For an anti-tumor immune response to be effective in eliminating cancer cells, a series of events must be initiated; these are collectively referred to as the cancer-immune cycle and include antigen release and presentation, immune cell priming and activation, T-cell trafficking and infiltration, and finally, recognition and killing of cancer cells [6]. lncRNAs expressed by tumor cells are involved in immune modulation mainly via regulation of antigen release and presentation, immune cell priming and activation, as well as reshaping the functions of immune cells.
Antigen release and presentation
Studies have shown that lncRNAs may accelerate tumor advancement by influencing the tumor antigens production and the MHC molecules expression. For example, ncRNA-RB1 serves as a tumor inhibitor in lung cancer by positively regulating the expression of calreticulin (CALR) [19], which is a calcium-binding chaperone in cancer cells that influences antigen presentation by facilitating the folding of MCH-I [20]. CALR is expressed on the cell surface and serves as a “kill me” signal that promotes phagocytosis of macrophages [21]. Knockdown of ncRNA-RB1 inhibits the expression of CALR and prevents the translocation of CALR to the cell surface, and this inhibits the cellular uptake of macrophages [19].
LncRNA inducing MHC-I and immunogenicity of tumor (LIMIT) is an immunogenic lncRNA found in humans and mice [22]. When induced by IFN-γ, LIMIT activates the guanylate-binding protein gene clusters, which release heat shock factor-1 (HSF1) from HSP90. This results in HSF1 activation and MHC-I transcription. Thus, LIMIT may be a potential epigenetic target for immunotherapy [22]. Overexpression of another lncRNA, LINC02195, induces the expression of MHC-I, thereby increasing the infiltration of T cells in head and neck cancer [23]. Further, another lncRNA, HOTAIR, can act as a ceRNA and induce HLA-G expression in gastric and cervical cancers, and HLA-G is known to be involved in tumor escape [24,25].
Priming and activation
Immune checkpoint molecules are crucial inhibitors in immune priming and activation. Research on how lncRNAs regulate this process mainly focuses on the modulation of programmed cell death 1 ligand 1 (PD-L1), the most widely used immune checkpoint inhibitor in the clinical setting [26]. In recent studies, lncRNAs were found to regulate the expression of PD-L1 by acting as ceRNAs that were capable of direct sponge adsorption of miRNAs in multiple cancers. In addition, the lncRNA UCA1 facilitates viability and metastatic potential of gastric carcinoma cells by targeting miRNAs, and may lead to immune evasion by upregulating PD-L1 expression [27]. In thyroid carcinoma, UCA1 induces PD-L1 expression and attenuates cytokine secretion in CD8+ T cells, thereby inhibiting the cytotoxic effect of CD8+ T cells and promoting cancer development [28]. Further, the lncRNA MALAT1 is a conserved lncRNA in mammals that can competitively bind with miRNAs to positively regulate the expression of PD-L1 in diffuse large B-cell lymphoma and NSCLC [29,30], facilitating apoptosis of CD8+ T cells [29]. Moreover, SNHG14 was found to increase the level of zinc finger E-box binding homeobox 1, which in turn, upregulates SNHG14 and PD-L1. Thus, the high expression of PD-L1 is maintained by a positive feedback loop, which inhibits the CTLs activation and leads to immune escape [31]. In addition, the lncRNA LINC00473 has also been found to stimulate PD-L1 expression in pancreatic cancer [32]. These findings indicate that lncRNAs promote the immune escape of cancer cells.
Of note, another lncRNA, lncMX1-215, is known to suppress tumor growth by negative regulation of PD-L1. When induced by IFNα, lncMX1-215 can significantly inhibit proliferation and migration of head and neck cancer cells, as well as downregulate IFNα-induced expression of PD-L1 and galectin-9. With regard to the underlying mechanism, the direct interaction between lncMX1-215 and GCN5, an H3K27 acetylase, prevents H3K27 acetylation on PD-L1 and galectin-9 promoters, thereby negatively regulating PD-L1 and galectin-9 expression and relieving immunosuppression [33] (Table 2).
|
Immune-related function |
LncRNA |
Functions and Mechanisms |
References |
|
Antigen release and presentation |
NcRNA-RB1 |
Promotes the expression and translocation of CALR, thus enhancing phagocytosis of macrophages |
[19] |
|
LncRNA LIMIT |
Activates HSF1 and the transcription of the MHC-I machinery |
[22] |
|
|
LINC02195 |
Upregulates the expression of MHC-I |
[23] |
|
|
Priming and activation |
LncRNA UCA1 |
Upregulates the expression of PD-L1 and inhibits the cytotoxic effect of CD8+ T cells |
[27] |
|
LncRNA MALAT1 |
Upregulates the expression of PD-L1 through ceRNA network |
[29,30] |
|
|
LncRNA SNHG14 |
Upregulates the expression of PD-L1 and forms a positive feedback loop |
[31] |
|
|
LncMX1-215 |
Downregulates the expression of PD-L1 by preventing H3K27 acetylation on PD-L1 promoters |
[33] |
|
|
Regulation of T cells |
LINC00301 |
Drives Treg infiltration by facilitating TGF-β1 secretion |
[36] |
|
LINC00240 |
Suppresses NKT cell activity by inhibiting the expression of MICA |
[37] |
|
|
Regulation of TAMs |
LncRNA XIST |
Recruits MDSCs and TAMs |
[39] |
|
Lnc-BM LncRNA LNMAT1 |
Induces TAMs migration by promoting expression of CCL2 |
[40,42] |
|
|
LncRNA RPPH1 LncRNA PCAT6 LINC00662 |
Boosts macrophage M2 polarization |
[44-46] |
|
|
LncRNA MALAT1 |
Prevents macrophage M1 polarization |
[47] |
|
|
Regulation of TANs |
LncRNA HOTTIP |
Induces PD-L1 expression of neutrophils and inhibits T cell proliferation by promoting IL-6 expression |
[48] |
|
LINC01116 |
Recruits TANs |
[49] |
|
|
Regulation of CAFs |
LncRNA POU3F3 lncRNA Gm26809 |
Mediates the reprogramming of normal fibroblasts into CAFs |
[50,51] |
Regulation of immune cells
Studies have suggested that there is a correlation between lncRNA expression and immune infiltration. For example, the lncRNA BM466146 upregulates CXCL13, which shows a positive association with enhanced T-cell infiltration in breast cancer [34]. Further, overexpression of the lncRNA NNT-AS1 activates the TGF-β signaling pathway in hepatocellular carcinoma (HCC), and this is correlated with a decrease in CD4+ lymphocyte infiltration [35]. Currently, there is ongoing research about how tumor cell-derived lncRNAs modulate immune cell function in the TME.
T cells: The lncRNA LINC00301, which is expressed at high levels in NSCLC, has been shown to facilitate TGF-β1 secretion to drive Treg cell infiltration and then decrease the CD8+ T-cell population in the TME [36]. In cervical cancer, LINC00240 induces STAT3 expression via the ceRNA mechanism. One of the downstream targets of STAT3 is MHC class I-related chain (MIC)-A, which is critical for natural killer T (NKT) cell activation. Accordingly, the expression of MICA was inhibited by LINC00240, and this led to the suppression of NKT cell activity [37].
Macrophages: The recruitment of macrophages is affected by some lncRNAs, according to recent evidence. In a pan-cancer RNA analysis, an lncRNA for calcium-dependent kinase activation (CamK-A) was identified as an oncogenic gene. CamK-A can trigger NF-κB activation by activating pregnancy up-regulated nonubiquitous CaM kinase (PNCK), which contributes to TME reshaping, including angiogenesis and macrophage recruitment [38]. A study on colitis-associated colorectal cancer has illustrated the crucial role of CXCR4 in exacerbating colitis-associated colorectal tumorigenesis and progression via the lncRNA XIST/miR-133a-3p/RhoA signaling pathway, which recruits MDSCs and macrophages [39]. In addition, a novel lncRNA associated with breast cancer brain metastasis, lnc-BM, was found to activate JAK2 kinase and mediate the phosphorylation of STAT3, thus promoting the STAT3-dependent expression of ICAM1 and CCL2. ICAM1 increases vascular cooption, while CCL2 induces macrophage migration into the brain by passing through the blood-brain barrier [40]. The recruited macrophages secrete oncostatin M and interleukin (IL)-6, which in turn further trigger STAT3 phosphorylation. This finding reveals a positive feedback loop that boosts breast cancer metastasis triggered by lnc-BM [41]. The macrophage recruitment function of lncRNA-induced CCL2 has also been reported in bladder cancer. lncRNA lymph node metastasis-associated transcript 1 or LNMAT1 was found to activate CCL2 expression by recruiting hnRNPL to the CCL2 promoter. The macrophages recruited by CCL2 promote lymphatic metastasis via VEGF-C excretion [42].
In general, macrophages can be classified as classically (M1 phenotype) or alternatively activated (M2 phenotype) macrophages according to the activation stage and functional status [43]. M1 macrophages are involved in the type I T helper cell (TH1) response against pathogens and cancer through the secretion of pro-inflammatory cytokines, while M2 macrophages are involved in wound repair and can induce tumorigenesis and immunosuppression in tumors [43]. Various reports on cancer have demonstrated the vital roles of lncRNAs in macrophage polarization. One report found that cancer cells deliver the lncRNA RPPH1 via exosomes to macrophages and boost M2 polarization, which consist of a positive feedback that enhances the viability and invasion of colorectal cancer cells [44]. In NSCLC, transport of PCAT6 from tumor cells was found to promote polarization of macrophages towards the M2 subset. Further, M2 macrophages were found to promote EMT and metastasis via the PCAT6/miR-326/KLF1 axis [45]. In HCC, LINC00662 was found to promote M2 macrophage polarization in a paracrine manner [46], and knockdown of the lncRNA MALAT1 was found to suppress angiogenesis and promote M1 macrophage polarization [47]. Thus, MALAT1 may act as a tumor-promoting marker in HCC.
Neutrophils: Tumor-associated neutrophils (TANs) have emerged as a critical component of the TME that play a role in tumor-related inflammation and immunosuppression. In ovarian cancer, the lncRNA HOTTIP, via upregulation of the secretion of IL-6, promotes PD-L1 expression on neutrophils by STAT3 phosphorylation, which consequently causes inhibition of T-cell proliferation and cancer cell escape [48]. In gliomas, LINC01116 activates IL-1β expression, contributing to the accumulation of TANs in the TME. The infiltrated TANs produce various cytokines to stimulate proliferation of cancer cells. Hence, the elevated expression of LINC01116 in glioma tissues is indicative of poor prognosis in patients [49].
Fibroblasts: Fibroblasts are stromal cells that can be activated in cancer and are commonly known as cancer-associated fibroblasts (CAFs). CAFs can interact with cancer cells to promote tumor metastasis and progression. Studies on how lncRNAs regulate fibroblasts to facilitate tumor advancement have illustrated that tumor cell-derived lncRNAs mainly regulate fibroblasts via exosomes. In esophageal squamous cell carcinoma, the lncRNA POU3F3, which is carried by exosomes produced by cancer cells, reprograms fibroblasts into CAFs. The activated fibroblasts secrete inflammatory cytokines, among which IL-6 promotes the proliferation and cisplatin resistance of esophageal squamous cell carcinoma (ESCC) cells [50]. In melanomas, tumor-secreted exosomal lncRNA Gm26809 was found to transform NIH/3T3 fibroblasts into CAFs, as evidenced by the increase in the expression of α-smooth actin and fibroblast activation protein. Coculture of CAFs and melanoma cells revealed significantly enhanced proliferation and migration of melanoma cells [51].
Overall, the findings described in this section indicate that lncRNAs derived from tumor cells are involved in various stages of the cancer-immune cycle in several types of cancers, as well as the regulation of immune cells. Thus, the mechanisms underlying their effects on the cancer-immune cycle may be related to their effects on the expression of specific proteins by immune cells and the functions of these cells.
In addition to cancer cells, other immune cells and stromal cells may also have an influence on anti-tumor activities via their effects on events of the cancer-immune cycle. Therefore, the next two sections focus on lncRNAs associated with these cells.
Immune Cell-derived lncRNAs
Modulation of immune cell differentiation
Various studies have reported that lncRNAs serve as crucial modulators in lymphoid and myeloid differentiation and activation [52-55]. Further, bioinformatics mining analyses have indicated that T cell-derived lncRNAs could affect the differentiation and function of T cells [56-58]. Zhang and his colleagues determined that the lncRNA lnc-DC interacts with STAT3 in the cytoplasm and activates the STAT3 pathway, affecting the differentiation and maturation of DCs [59]. Further, research by Jonathan and his colleagues has revealed that the lncRNA Morrbid regulates the lifespan of myeloid cells by maintaining the pro-apoptotic gene Bcl2l11 [60]. Furthermore, long noncoding monocytic RNA (lnc-MC) was found to enhance the expression of activin A receptor type 1B, which promotes activation of the TGF-β signaling pathway, thus ultimately promoting the differentiation of monocytes and macrophages [61].
Researchers have characterized lncRNAs in immune cell differentiation and development, and recently, more studies have been exploring how these lncRNAs promote tumor progression or suppress tumor growth. In one such study, overexpression of lnc-epidermal growth factor receptor (EGFR) in Tregs was found to correlate positively with the expression of EGFR/Foxp3 in patients with HCC. lnc-EGFR targets EGFR and blocks its interaction and ubiquitination via c-CBL. Stabilization of EGFR enables activation of the AP-1/NF-AT1 pathway, which in turn enhances the expression of EGFR. Modulation of EGFR by lnc-EGFR results in Treg differentiation and CTL suppression, and this promotes tumor growth and immune escape [62]. It has been reported that another lncRNA, namely SNHG1, plays a vital role in modulating Treg differentiation. In breast cancer, blocking SNHG1 has been found to downregulate IDO expression and inhibit Treg differentiation, thereby suppressing tumor growth [12] (Table 3).
|
Immune-related function |
LncRNA |
Functions and Mechanisms |
References |
|
Modulation of immune cell differentiation |
Lnc-EGFR |
Modulates Treg differentiation and CTL suppression |
[62] |
|
LncRNA SNHG1 |
Promotes Treg differentiation by upregulation of IDO |
[12] |
|
|
Antigen presentation |
Lnc-DC |
Impairs the antigen uptake of DCs by downregulating the expression of HLA-DR |
[64] |
|
Lnc-Dpf3 |
Targets HIF-1α and suppresses Ldha |
[65] |
|
|
Regulation of CD8+T cells |
Lnc-Tim3 |
Maintains exhausted CD8+ T cells by inducing expression of the anti-apoptotic proteins |
[66] |
|
LncRNA NEAT1 |
Promotes CD8+ T cell apoptosis through miR-155/Tim-3 pathway |
[15] |
|
|
Regulation of other immune cells |
LncRNA-MM2P |
Promotes macrophage M2 polarization |
[67] |
|
LncRNA NIFK-AS1 LncRNA Cox-2 |
Inhibits macrophage M2 polarization |
[68,69] |
|
|
LincRNA-p21 LncRNA TUC339 LncRNA ANCR |
Inhibits macrophage M1 polarization |
[70-72] |
|
|
LncRNA Pvt1 |
Inhibits MDSCs by upregulating HIF-1a |
[74] |
|
|
Regulation of tumor cells |
LncRNA HISLA |
Enhances the aerobic glycolysis by stabilizing HIF-1a |
[77] |
|
LncRNA AFAP1-AS1 |
Promotes migration and invasion by inhibiting miR-26a expression and upregulating ATF2 |
[78] |
Antigen presentation
Dendritic cells are the prominent antigen-presenting cells in the TME. After phagocytosis of antigens, they mature under cytokine stimulation, express co-stimulatory molecules such as CD80/86/40, and present antigens to T cells to activate immune responses [63]. Silencing of lnc-DC has been found to suppress the expression of HLA-DR and impair the antigen uptake of DCs, thereby reducing the ability of DCs to boost T-cell proliferation. In other words, lnc-DC can enhance T-cell priming and inflammatory cytokine secretion [64].
DC trafficking is also required to initiate immune defense. Chemokine receptor type 7 (CCR7) ligands have been demonstrated to be important in the migration of DCs. CCR7 stimulation promotes lnc-Dpf3 expression by preventing its degradation, and CCR7 induces metabolic reprogramming toward glycolysis. This promotes DC migration by activating the HIF-1α pathway in DCs. However, lnc-Dpf3 can directly target HIF-1α and suppress Ldha, a glycolytic gene that is transcribed in an HIF-1α-dependent manner. This results in the constitution of a negative feedback loop that inhibits the glycolytic metabolism and migratory capacity of DCs, and causes termination of CCR7-mediated DC migration and helps to maintain immune homeostasis [65] (Table 3).
Modulation of the cytotoxic effect of CD8+ T cells
Currently, lncRNAs are considered as important regulators of CD8+ T cell function. For example, Ji and his colleagues discovered that lnc-Tim3 is a pivotal regulator of CD8+ T cell exhaustion and survival. lnc-Tim3 is highly expressed by CD8+ T cells in HCC, and its expression correlates negatively with IFN-γ and IL-2 production. These findings suggest that lnc-Tim3 promotes CD8+ T cell exhaustion. An in-depth study into this effect revealed that lnc-Tim3 could inhibit the interaction between Tim-3 and Bat3 by specifically binding to Tim-3, thus resulting in the nuclear localization of Bat3. Furthermore, lnc-Tim3 was found to enhance the transcriptional activation of p53 and RelA and induce expression of the anti-apoptotic proteins p21, MDM2, and Bcl-2, thus leading to the survival of Tim-3+ exhausted CD8+ T cells [66]. In another study exploring the roles of CD8+ T cell-derived lncRNAs during HCC development, the lncRNA NEAT1 was found to participate in immune surveillance escape of HCC through the miR-155/Tim-3 pathway in CD8+ T cells, thus promoting CD8+ T-cell apoptosis and suppressing cytolysis [15] (Table 3).
Modulation of other immune cells
Based on the accumulating evidence for the pivotal role of lncRNAs in modulating immune cell differentiation and function, Cao and his colleagues tried to identify the lncRNAs of macrophages that modulate macrophage polarization. Using lncRNA arrays, they determined that lncRNA-MM2P serves as a modulator of macrophage M2 polarization. That is, inhibition of lncRNA-MM2P was found to inhibit macrophage polarization to M2 and reduce angiogenesis of M2 by blocking STAT6 phosphorylation [67]. On the contrary, the lncRNAs NIFK-AS1 and Cox-2 were found to suppress M2 polarization of macrophages [68,69]. In addition, knockdown of lncRNAs, such as lincRNA-p21, lncRNA TUC339, and lncRNA ANCR, could promote macrophage polarization into pro-inflammatory M1 subsets [70-72] (Table 3).
MDSCs, which are pathologically activated immature cells, participate in tumor-elicited immunosuppression. These cells dramatically suppress the T cell-induced anticancer response, thereby contributing to the immune escape of malignant cells [73]. The lncRNA Pvt1 has been revealed as a suppressor in the functional regulation of MDSCs. Under hypoxic stress, Pvt1 is upregulated by HIF-1a in MDSCs, and this remarkably inhibits the function of MDSCs. Accordingly, Pvt1 knockdown was found to reduce the immunosuppressive ability of MDSCs, thereby accelerating tumor progression and inhibiting anti-tumor immune responses [74]. Other studies have demonstrated that some lncRNAs, for instance, Lnc-C/EBPβ and Lnc-chop, inhibit the immune suppressive function of MDSCs by regulating target transcripts, including arginase-1, nitric oxide synthase 2, NADPH oxidase 2, and cyclooxygenase-2, all of which are closely related to the immunosuppressive function of MDSCs in the TME [75,76].
Modulation of tumor cells via extracellular vesicles
LncRNAs have been found to be transported from immune cells to tumor cells through extracellular vesicles. For example, the lncRNA HIF-1α-stabilizing long non-coding RNA (HISLA), which is transmitted by extracellular vesicles from tumor associated macrophages (TAMs) to breast cancer cells, enhances the aerobic glycolysis of cancer cells by blocking the binding of PHD2 and HIF-1α and stabilizing HIF-1α. Correspondingly, glycolytic tumor cells deliver lactic acid to further facilitate HISLA expression in TAMs, connecting TAMs and tumor cells in a feed-forward loop [77]. Another example is that of the lncRNA AFAP1-AS1 from M2 macrophage-derived exosomes, which can inhibit the expression of miR-26a and upregulate ATF2, consequently promoting the migration, invasion, and lung metastasis of esophageal cancer cells [78] (Table 3).
Overall, the findings so far demonstrate that several lncRNAs derived from immune cells are closely involved in the modulation of tumor cells and various types of immune cells. LncRNAs expressed in the immune cells, such as CD8+ T cells and MDSCs, participate in regulating their functions, such as differentiation, antigen presentation, and cytotoxic effects. Yet, more research is need to further illustrate how immune cell-derived lncRNAs affect another type of immune cell.
Stromal Cell-derived lncRNAs
Stromal CAFs are the most important stromal components in the TME [79]. Ding and his colleagues uncovered a so-far uncharacterized lncRNA, FLJ22447, which is remarkably upregulated during the transformation of normal fibroblasts to CAFs, and they named it lnc-CAF. With regard to its mechanism of action, lnc-CAF maintains the proliferative effect of IL-33 on tumor cells by directly upregulating IL-33 and preventing degradation of IL-33 by p62-dependent autophagy-lysosome. In turn, tumor cells further induce an increase in lnc-CAF levels in stromal fibroblasts via exosomal lnc-CAF [80] (Table 4).
|
Regulation of tumor cells |
LncRNA |
Functions and Mechanisms |
References |
|
Promotion the aggressiveness of tumor cells |
Lnc-CAF |
Upregulates IL-33 and maintains effects of IL-33 on tumor proliferation |
[80] |
|
LncRNA H19 LncRNA CCAL |
Promotes aggressiveness and chemoresistance by activating the β-catenin pathway |
[81,82] |
|
|
Reprograms of the metabolic pathways |
LINC01614 |
Enhances glutamine uptake by directly interacting with ANXA2 and p65 to facilitate the activation of NF-κB |
[84] |
|
LncRNA SNHG3 |
Inhibits mitochondrial oxidative phosphorylation and increases glycolysis and carboxylation, thus enhancing tumor proliferation |
[85] |
Exosomes from CAFs, carrying lncRNAs, can participate in tumor pathogenesis and progression by modulating the behavior of tumor cells. In colorectal cancer, the lncRNA H19 and colorectal cancer-associated lncRNA (CCAL) are more abundant in the stroma than in the tumor nests. They are transported to cancer cells from CAF-secreted exosomes. Both lncRNA H19 and CCAL can promote aggressiveness and chemoresistance by activating the β-catenin pathway [81,82]. Similarly, lncRNA TIRY-overexpressing CAF-derived exosomes deliver miR-14 to oral squamous cell carcinoma (OSCC) cells, and this promotes cancer cell invasion and metastasis [83].
In addition to enhancing aggressiveness, lncRNAs from CAF-derived exosomes were also able to reprogram the metabolic pathways of cancer cells. For example, a CAF-specific lncRNA, LINC01614, was found to strengthen glutamine uptake in lung adenocarcinoma (LUAD) cells [84]. In addition, the lncRNA SNHG3 can inhibit mitochondrial oxidative phosphorylation and increase glycolysis and carboxylation after uptake of exosomes by tumor cells, and this leads to enhanced tumor cell proliferation [85] (Table 4). Thus, CAFs seem to be the prominent source of lncRNAs derived from the stroma that are associated with cancer proliferation and metastasis.
Other than fibroblasts, lncRNAs also have impact on the function of endothelial cells. Researchers have discovered lncRNA n342419, which they termed it MANTIS, serves as an endothelial angiogenic facilitator [86]. Moreover, lncRNA-MIAT can ameliorate diabetes mellitus-induced retinal microvascular dysfunction by forming a feedback loop with vascular endothelial growth factor and miR-150-5p [87]. However, more efforts to demonstrate how lncRNAs function in cancer associated endothelial cells are still in urgent need.
The Clinical Implication of Immune-related lncRNAs
Therapeutic potential of immune-related lncRNAs
From a clinical perspective, lncRNAs may serve as promising targets for tumor therapy since they extensively mediate tumor progression. Currently, there have been significant advances in technologies targeting lncRNAs [88], including techniques to modulate nuclear and cytoplasmic lncRNA expression: (1) Blocking lncRNA transcription through the integration of RNA destabilizing elements (RDE) into the genomic locus [89]. (2) Destabilizing lncRNA transcript by siRNA [3], antisense oligonucleotides (ASO) [90] and locked nucleic acid (LNA) GapmeRs [91]. (3) Inhibiting interactions of lncRNAs with others by small molecules and aptamer [92]. (4) Gene editing for lncRNAs using CRISPR [93].
These approaches have been validated in patient-derived tumor xenograft (PDX) models. Researchers have demonstrated that siRNA targeting lnc-BM can inhibit brain metastasis of breast cancer. Lnc-BM induces the expression of ICAM1 and CCL2, resulting in macrophage chemotaxis into the brain, and eventually assists brain metastasis. To explore a potential therapeutic strategy against the disease, researchers developed brain metastasis-bearing mice models and delivered nanoparticle-conjugated siRNAs for lnc-BM. In vivo experiments confirmed that depletion of lnc-BM by siRNAs effectively suppressed brain metastasis [41]. In addition, targeting lncRNA may also improve the effectiveness of immunotherapy. The increasing level of LIMIT by CRISPR activation can promote antigen presentation by boosting MHC-I expression, thereby exhibiting a synergistic anti-tumor effect with immune checkpoint blockade (ICB) [22]. In immunocompromised mice, NKILA silencing in T cells restrains them from immunological elimination, which enhances the efficacy of adoptive T cell therapy [18]. These findings suggest the therapeutic potential of lncRNAs in preclinical models.
LncRNA-related therapeutic strategies have several advantages. First, considering some dysregulated lncRNAs are cancer-specific, these lncRNAs are critical for the development of personalized therapy. Also, lncRNAs interact with other molecules on multiple regulatory sites, providing more opportunities for novel structure-based drug development [94].
Although targeting lncRNAs displays therapeutic potential, some issues in translating into the clinic including delivery, immunogenicity and specificity, are similar to all RNA-based treatments [95]. In terms of delivery and immunogenicity, one solution is to encapsulate lncRNAs or lncRNA-targeting molecules with extracellular vesicles [96], which are immune tolerant and tissue penetrating. An alternative is to use artificial carriers, for example, synthetic nanoparticles [41]. To improve the specificity, modification of the RNAs or nanoparticles may strengthen the on-target specificity. However, it remains uncertain whether any deviation from the target would pose a safety risk. To date, several mRNA-based therapeutics, either siRNAs or ASOs, have gained FDA approval, but no lncRNA-based ones have been applied in the clinic [95]. The research of lncRNAs on tumor therapy is mostly based on mice models, and there is still a long way to go before lncRNAs or lncRNA-targeted molecules can be used in tumor treatment.
The prognostic value of immune-related lncRNAs
LncRNAs regulate crucial mechanisms of cancer immunity, and this might imply that they have prognostic potential [5,36,41,48,50,80]. In addition to individual lncRNAs, a panel of lncRNAs can also be used for prognosis prediction in cancer patients. Inspired by studies on the metastasis-promoting roles of lncRNAs in nasopharyngeal carcinoma (NPC) patients [97-100], we conducted a retrospective multicohort study with the aim of developing an lncRNA signature for NPC metastasis prediction and exploring its potential function [101]. We employed a three-step strategy to develop a lncRNA signature to predict metastasis, including screening of lncRNAs by microarrays in matched samples in a discovery cohort, model training with a larger sample size in the training cohort, and validation in two independent cohorts. The nine-lncRNA signature (comprising lnc-TRAPPC6B-2, lnc-DRD5-10, NR2F2-AS1, lnc-CETP-1, lnc-CDK1-1, LINC02065, lnc-POTEH-7, lnc-STX6-2, and lnc-C11orf91-2) could be used to reliably classify NPC patients according to the risk of distant metastasis. Surprisingly, we discovered that the lncRNAs included in our signature were associated with immune features. In addition, digital pathology studies also confirmed the difference in the degree of immune infiltration between high-risk and low-risk groups, as the low-risk population was found to have a higher population of CD8+ T cells and B cells in both tumor nests and stroma. Overall, our results suggest that immune-related lncRNAs play an essential role in the metastasis of NPC [101].
Unlike our previous study, other studies start with the identification of immune-related lncRNAs, either by correlation analysis between lncRNAs and corresponding mRNAs [102-104] or by recognition of specific immune cell-derived lncRNAs [105,106], and then explore their potential prognostic value. Immune-related lncRNA signatures identified in such studies have been found to be robust for survival prediction in HCC, bladder cancer, and colorectal cancer [103,104,107]. In addition, a pan-cancer analysis has also characterized immune-related lncRNAs as oncogenic biomarkers [102].
The tumor-infiltrating immune-related lncRNA signature (TILSig) is a collection of seven lncRNAs that are specifically expressed in immune cell lines rather than NSCLC cell lines: HCG26, PSMB8-AS1, TNRC6C-AS1, CARD8-AS1, HCP5, LOC286437, and LINC02256. Expression of the seven lncRNAs individually is associated with the overall survival of NSCLC patients. Further, the combination of the seven lncRNAs weighted by their coefficients according to multivariate Cox analysis can be used to categorize NSCLC patients into high-risk and low-risk subsets with significantly different clinical outcomes. The low-risk patients have a higher degree of immune cell infiltration: that is, the tumors in these patients are characterized by a higher number of activated CD8+ T cells and activated DCs. In contrast, the high-risk patients have a higher degree of infiltration of activated CD4+ T cells. TILSig is also an independent prognostic factor for overall survival prediction and serves as an indicator of immunotherapy response [105]. Similarly, TILBlncSig is an eight-lncRNA panel (comprising TNRC6C-AS1, WASIR2, GUSBP11, OGFRP1, AC090515.2, PART1, MAFG-DT and LINC01184) that represents infiltration of B lymphocytes in bladder cancer; it is comprised of lncRNAs exclusively expressed in B cells and can be used to discriminate between patients with disparate clinical outcomes in multicohort studies [106].
Prospective
As research on lncRNAs has become more intensive, an increasing number of novel lncRNAs are being discovered. In the future, more investigations are required to determine the functions of these novel lncRNAs. To date, studies on lncRNAs have relied heavily on microarray detection or bulk RNA sequencing, which limits the exploration of cell-type specific lncRNA functions in the TME. Some investigators have conducted studies on specific isolated cell types or cell lines, with the aim of determining the role and prognostic value of lncRNAs in specific immune cells [64,66,105,106,108]. Although these studies have yielded some remarkable findings, the contributions of individual cell types to the expression of these lncRNAs still need to be investigated. Single-cell RNA sequencing (scRNA-seq) serves as a powerful solution to analyze the expression of lncRNAs in a single cell. It may contribute to the discovery of new cell subtypes defined by lncRNA.
Recently, single-cell analyses have allowed for further discovery of new lncRNA markers and their functions in embryo and stem cell development [109-111], as well as viral infectious diseases [112-115]. Furthermore, studies on lncRNAs at the single-cell level have been started in the field of cancer research. For example, data on scRNA-seq of clear cell renal cell carcinoma (ccRCC) have revealed 173 ccRCC metastasis-associated lncRNAs that contribute to cell adhesion, proliferation, and immune response [116]. In addition, an M2 macrophage polarization-associated lncRNA was screened out by scRNA-seq in HCC, and found to promote glucose metabolism and cell proliferation by acting as miRNA sponge [117]. However, the number of studies on lncRNAs using single-cell analyses are still limited, partly due to the lack of annotations for new lncRNAs and failure to detect low-abundance transcripts.
Sequencing technologies have led to an “omics” revolution, with large information datasets. However, how to interpret these data to further help us understand the role of immune-related lncRNAs remains a considerable challenge. Artificial intelligence (AI) can serve as an ideal tool by accurately interpreting omics data using machine learning and deep learning, as well as integrating data from medical and pathological images [118]. Liu and his colleagues develop a machine learning-based integrative procedure for constructing a consensus immune-related lncRNA signature (IRLS), which has robust predictive value for colorectal cancer [107]. In our previous study, a machine learning-based LASSO algorithm [119] was used to develop a lncRNA-based prognostic signature, and digital pathology consisting of region annotation [120], image segmentation [121-123] and positively-staining cell identification [124] was used to evaluate immune infiltration in the sub-groups divided according to the signature [101].
Conclusion
This review summarizes the roles of lncRNAs expressed by various cells that regulate cancer immunity in the TME. Immune-related RNAs are involved in immunoregulatory processes and can be expressed by immune cells, tumor cells, or stromal cells. lncRNAs of different origins can affect the behavior of other cell types through the paracrine system or extracellular vesicles, and this implies that regulation of the TME is not an isolated intracellular process, but rather, it involves subtle interactions among various cell types. Yet, how lncRNAs interact with other noncoding RNAs in regulating the TME is not discussed here in detail. Instead, we also discussed potential therapeutic strategies based on lncRNAs and their drawbacks. Although investigations into the expression and characterization of lncRNAs currently rely on bulk RNA sequencing, we believe that rapid advances in single-cell methods and bioinformatics analytical tools will tremendously help to further illustrate the regulatory roles and clinical value of immune-related lncRNAs [125].
Conflicts of Interest
All authors declare no conflict of interest.
Funding Statement
This study did not receive any funds.
Abbreviations
lncRNAs: Long non-coding RNAs; TME: Tumor Microenvironment; DC: Dendritic Cell; MHCI: Major Histocompatibility Class I; Treg: Regulatory T cell; MDSC: Myeloid-Derived Suppressor Cell; EMT: Epithelial-Mesenchymal Transition; NSCLC: Non-Small-Cell Lung Cancer; VEGF: Vascular Epithelial Growth Factor; ceRNA: Competing endogenous RNA; SNHG1: Small Nucleolar RNA Host Gene 1; IDO: Indoleamine 2,3-dioxygenase; NEAT1: Nuclear Enriched Abundant Transcript 1; NKILA: NF-κB-Interacting lncRNA; CTLs: Cytotoxic T Lymphocytes; CALR: Calreticulin; LIMIT: LncRNA Inducing MHC-I and Immunogenicity of Tumor; HSF1: Heat Shock Factor-1; PD-L1: Programmed cell death 1 Ligand 1; HCC: Hepatocellular Carcinoma; MIC: MHC class I-related Chain; NKT: Natural Killer T; CamK-A: Calcium-dependent Kinase Activation; PNCK: Pregnancy up-regulated Nonubiquitous CaM Kinase; TH1: Type I T Helper cell; TANs: Tumor-Associated Neutrophils; CAFs: Cancer-Associated Fibroblasts; ESCC: Esophageal Squamous Cell Carcinoma; lnc-MC: Long noncoding Monocytic RNA; EGFR: Epidermal Growth Factor Receptor; CCR7: Chemokine Receptor type 7; HISLA: HIF-1α-Stabilizing Long non-coding RNA; TAMs: Tumor Associated Macrophages; CCAL: Colorectal Cancer-Associated lncRNA; OSCC: Oral Squamous Cell Carcinoma; LUAD: Lung Adenocarcinoma; RDE: RNA Destabilization Elements; ASO: Antisense Oligonucleotide; LNA: Locked Nucleic Acid; ICB: Immune Checkpoint Inhibitor; PDX: Patient-Derived tumor Xenograft; NPC: Nasopharyngeal Carcinoma; TILSig: Tumor-infiltrating immune-related lncRNA signature; scRNA-seq: Single-cell RNA sequencing; ccRCC: Clear cell Renal Cell Carcinoma; AI: Artificial Intelligence; IRLS: Immune-Related lncRNA Signature
References
2. Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337(6099):1159-61.
3. Zheng ZQ, Li ZX, Guan JL, Liu X, Li JY, Chen Y, et al. Long Noncoding RNA TINCR-Mediated Regulation of Acetyl-CoA Metabolism Promotes Nasopharyngeal Carcinoma Progression and ChemoresistanceTINCR Promotes Cancer Progression and Chemoresistance. Cancer Research. 2020 Dec 1;80(23):5174-88.
4. Zhang H, Hua Y, Jiang Z, Yue J, Shi M, Zhen X, et al. Cancer-associated fibroblast-promoted lncRNA DNM3OS confers radioresistance by regulating DNA damage response in esophageal squamous cell carcinoma. Clinical Cancer Research. 2019 Mar 15;25(6):1989-2000.
5. Zhao L, Ji G, Le X, Wang C, Xu L, Feng M, et al. Long noncoding RNA LINC00092 acts in cancer-associated fibroblasts to drive glycolysis and progression of ovarian cancer. Cancer Research. 2017 Mar 15;77(6):1369-82.
6. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013 Jul 25;39(1):1-0.
7. Shi X, Liu Z, Liu Z, Feng X, Hua F, Hu X, et al. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018 Nov 1;37:177-87.
8. Lang C, Yin C, Lin K, Li Y, Yang Q, Wu Z, et al. m6A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2‐mediated IGF1R mRNA stabilization. Clinical and Translational Medicine. 2021 Jun;11(6):e426.
9. Dong F, Ruan S, Wang J, Xia Y, Le K, Xiao X, Hu T, Wang Q. M2 macrophage-induced lncRNA PCAT6 facilitates tumorigenesis and angiogenesis of triple-negative breast cancer through modulation of VEGFR2. Cell death & disease. 2020 Sep 9;11(9):728.
10. Tu J, Wu F, Chen L, Zheng L, Yang Y, Ying X, et al. Long non-coding RNA PCAT6 induces M2 polarization of macrophages in cholangiocarcinoma via modulating miR-326 and RhoA-ROCK signaling pathway. Frontiers in Oncology. 2021 Jan 21;10:605877.
11. Hu Y, Ma Z, He Y, Liu W, Su Y, Tang Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochemical and Biophysical Research Communications. 2017 Sep 30;491(4):926-31.
12. Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. International Journal of Biological Macromolecules. 2018 Oct 15;118:24-30.
13. Zhang J, Li Y, Dong M, Wu D. Long non coding RNA NEAT1 regulates E2F3 expression by competitively binding to miR 377 in non small cell lung cancer. Oncology Letters. 2017 Oct 1;14(4):4983-8.
14. Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nature Communications. 2014 Nov 21;5(1):5383.
15. Yan K, Fu Y, Zhu N, Wang Z, Hong JL, Li Y, et al. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8+ T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. The International Journal of Biochemistry & Cell Biology. 2019 May 1;110:1-8.
16. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015 Mar 9;27(3):370-81.
17. Wu W, Chen F, Cui X, Yang L, Chen J, Zhao J, et al. LncRNA NKILA suppresses TGF‐β‐induced epithelial–mesenchymal transition by blocking NF‐κB signaling in breast cancer. International Journal of Cancer. 2018 Nov 1;143(9):2213-24.
18. Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nature Immunology. 2018 Oct;19(10):1112-25.
19. Musahl AS, Huang X, Rusakiewicz S, Ntini E, Marsico A, Kroemer G, et al. A long non-coding RNA links calreticulin-mediated immunogenic cell removal to RB1 transcription. Oncogene. 2015 Sep;34(39):5046-54.
20. Raghavan M, Wijeyesakere SJ, Peters LR, Del Cid N. Calreticulin in the immune system: ins and outs. Trends in Immunology. 2013 Jan 1;34(1):13-21.
21. Chao MP, Jaiswal S, Weissman-Tsukamoto R, Alizadeh AA, Gentles AJ, Volkmer J, et al. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Science Translational Medicine. 2010 Dec 22;2(63):63ra94.
22. Li G, Kryczek I, Nam J, Li X, Li S, Li J, et al. LIMIT is an immunogenic lncRNA in cancer immunity and immunotherapy. Nature Cell Biology. 2021 May;23(5):526-37.
23. Li H, Xiong HG, Xiao Y, Yang QC, Yang SC, Tang HC, et al. Long non-coding RNA LINC02195 as a regulator of MHC I molecules and favorable prognostic marker for head and neck squamous cell carcinoma. Frontiers in Oncology. 2020 May 6;10:615.
24. Song B, Guan Z, Liu F, Sun D, Wang K, Qu H. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochemical and Biophysical Research Communications. 2015 Aug 28;464(3):807-13.
25. Sun J, Chu H, Ji J, Huo G, Song Q, Zhang X. Long non-coding RNA HOTAIR modulates HLA-G expression by absorbing miR-148a in human cervical cancer. International Journal of Oncology. 2016 Sep 1;49(3):943-52.
26. Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nature reviews Clinical Oncology. 2021 Jun;18(6):345-62.
27. Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q, et al. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Molecular Cancer. 2019 Dec;18(1):115.
28. Wang X, Zhang Y, Zheng J, Yao C, Lu X. LncRNA UCA1 attenuated the killing effect of cytotoxic CD8+ T cells on anaplastic thyroid carcinoma via miR-148a/PD-L1 pathway. Cancer Immunology, Immunotherapy. 2021 Aug;70:2235-45.
29. Wang QM, Lian GY, Song Y, Huang YF, Gong Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sciences. 2019 Aug 15;231:116335.
30. Wei S, Wang K, Huang X, Zhao Z, Zhao Z. LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. International Journal of Immunopathology and Pharmacology. 2019 Jun;33:2058738419859699.
31. Zhao L, Liu Y, Zhang J, Liu Y, Qi Q. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death & Disease. 2019 Sep 30;10(10):731.
32. Zhou WY, Zhang MM, Liu C, Kang Y, Wang JO, Yang XH. Long noncoding RNA LINC00473 drives the progression of pancreatic cancer via upregulating programmed death‐ligand 1 by sponging microRNA‐195‐5p. Journal of Cellular Physiology. 2019 Dec;234(12):23176-89.
33. Ma H, Chang H, Yang W, Lu Y, Hu J, Jin S. A novel IFNα-induced long noncoding RNA negatively regulates immunosuppression by interrupting H3K27 acetylation in head and neck squamous cell carcinoma. Molecular Cancer. 2020 Dec;19:1-6.
34. Zhang Y, Dong X, Wang Y, Wang L, Han G, Jin L, et al. Overexpression of LncRNA BM466146 predicts better prognosis of breast cancer. Frontiers in Oncology. 2021 Jan 29;10:628757.
35. Wang Y, Yang L, Dong X, Yang X, Zhang X, Liu Z, et al. Overexpression of NNT-AS1 activates TGF-β signaling to decrease tumor CD4 lymphocyte infiltration in hepatocellular carcinoma. BioMed Research International. 2020 Dec 24;2020:8216541.
36. Sun CC, Zhu W, Li SJ, Hu W, Zhang J, Zhuo Y, Zhang H, Wang J, Zhang Y, Huang SX, He QQ. FOXC1-mediated LINC00301 facilitates tumor progression and triggers an immune-suppressing microenvironment in non-small cell lung cancer by regulating the HIF1α pathway. Genome Medicine. 2020 Dec;12(1):77.
37. Zhang Y, Li X, Zhang J, Liang H. Natural killer T cell cytotoxic activity in cervical cancer is facilitated by the LINC00240/microRNA-124-3p/STAT3/MICA axis. Cancer Letters. 2020 Apr 1;474:63-73.
38. Sang LJ, Ju HQ, Liu GP, Tian T, Ma GL, Lu YX, et al. LncRNA CamK-A regulates Ca2+-signaling-mediated tumor microenvironment remodeling. Molecular Cell. 2018 Oct 4;72(1):71-83.e7.
39. Yu X, Wang D, Wang X, Sun S, Zhang Y, Wang S, et al. CXCL12/CXCR4 promotes inflammation-driven colorectal cancer progression through activation of RhoA signaling by sponging miR-133a-3p. Journal of Experimental & Clinical Cancer Research. 2019 Dec;38:32.
40. Singh AV, Chandrasekar V, Laux P, Luch A, Dakua SP, Zamboni P, et al. Micropatterned Neurovascular Interface to Mimic the Blood-Brain Barrier's Neurophysiology and Micromechanical Function: A BBB-on-CHIP Model. Cells. 2022 Sep 8;11(18):2801.
41. Wang S, Liang K, Hu Q, Li P, Song J, Yang Y, Yao J, Mangala LS, Li C, Yang W, Park PK. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. The Journal of Clinical Investigation. 2017 Dec 1;127(12):4498-515.
42. Chen C, He W, Huang J, Wang B, Li H, Cai Q, et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nature Communications. 2018 Sep 20;9(1):3826.
43. Mohapatra S, Pioppini C, Ozpolat B, Calin GA. Non-coding RNAs regulation of macrophage polarization in cancer. Molecular Cancer. 2021 Dec;20:24.
44. Liang ZX, Liu HS, Wang FW, Xiong L, Zhou C, Hu T, et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death & Disease. 2019 Nov 4;10(11):829.
45. Chen Y, Hong C, Qu J, Chen J, Qin Z. Knockdown of lncRNA PCAT6 suppresses the growth of non-small cell lung cancer cells by inhibiting macrophages M2 polarization via miR-326/KLF1 axis. Bioengineered. 2022 May 2;13(5):12834-46.
46. Tian X, Wu Y, Yang Y, Wang J, Niu M, Gao S, et al. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/β-catenin signaling. Molecular Oncology. 2020 Feb;14(2):462-83.
47. Hou ZH, Xu XW, Fu XY, Zhou LD, Liu SP, Tan DM. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. American Journal of Physiology-Cell Physiology. 2020 Mar 1;318(3):C649-63.
48. Shang A, Wang W, Gu C, Chen C, Zeng B, Yang Y, et al. Long non-coding RNA HOTTIP enhances IL-6 expression to potentiate immune escape of ovarian cancer cells by upregulating the expression of PD-L1 in neutrophils. Journal of Experimental & Clinical Cancer Research. 2019 Dec;38(1):411.
49. Wang T, Cao L, Dong X, Wu F, De W, Huang L, et al. LINC01116 promotes tumor proliferation and neutrophil recruitment via DDX5-mediated regulation of IL-1β in glioma cell. Cell Death & Disease. 2020 May 1;11(5):302.
50. Tong Y, Yang L, Yu C, Zhu W, Zhou X, Xiong Y, et al. Tumor-secreted exosomal lncRNA POU3F3 promotes cisplatin resistance in ESCC by inducing fibroblast differentiation into CAFs. Molecular Therapy-Oncolytics. 2020 Sep 25;18:1-3.
51. Hu T, Hu J. Melanoma-derived exosomes induce reprogramming fibroblasts into cancer-associated fibroblasts via Gm26809 delivery. Cell Cycle. 2019 Nov 17;18(22):3085-94.
52. Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. The Journal of Immunology. 2012 Sep 1;189(5):2084-8.
53. Gao Y, Shang W, Zhang D, Zhang S, Zhang X, Zhang Y, et al. Lnc-C/EBPβ modulates differentiation of MDSCs through downregulating IL4i1 with C/EBPβ LIP and WDR5. Frontiers in Immunology. 2019 Jul 17;10:1661.
54. Tian X, Zheng Y, Yin K, Ma J, Tian J, Zhang Y, et al. LncRNA AK036396 inhibits maturation and accelerates immunosuppression of polymorphonuclear myeloid-derived suppressor cells by enhancing the stability of ficolin BLncRNA AK036396/Fcnb accelerates PMN-MDSC immunosuppression. Cancer Immunology Research. 2020 Apr 1;8(4):565-77.
55. Shang W, Gao Y, Tang Z, Zhang Y, Yang R. The Pseudogene Olfr29-ps1 Promotes the Suppressive Function and Differentiation of Monocytic MDSCsOlfr29-ps1 Regulates MDSC Differentiation. Cancer Immunology Research. 2019 May 1;7(5):813-27.
56. Kanduri K, Tripathi S, Larjo A, Mannerström H, Ullah U, Lund R, et al. Identification of global regulators of T-helper cell lineage specification. Genome Medicine. 2015 Dec;7:122.
57. Panzeri I, Rossetti G, Abrignani S, Pagani M. Long intergenic non-coding RNAs: Novel drivers of human lymphocyte differentiation. Frontiers in Immunology. 2015 Apr 15;6:175.
58. Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. The Journal of Immunology. 2009 Jun 15;182(12):7738-48.
59. Zhang W, Yang M, Yu L, Hu Y, Deng Y, Liu Y, et al. Long non‐coding RNA lnc‐DC in dendritic cells regulates trophoblast invasion via p‐STAT3‐mediated TIMP/MMP expression. American Journal of Reproductive Immunology. 2020 Jun;83(6):e13239.
60. Kotzin JJ, Spencer SP, McCright SJ, Kumar DB, Collet MA, Mowel WK, et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. 2016 Sep 8;537(7619):239-43.
61. Chen MT, Lin HS, Shen C, Ma YN, Wang F, Zhao HL, et al. PU. 1-regulated long noncoding RNA lnc-MC controls human monocyte/macrophage differentiation through interaction with microRNA 199a-5p. Molecular and Cellular Biology. 2015 Sep 15;35(18):3212-24.
62. Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nature Communications. 2017 May 25;8(1):15129.
63. Liu YJ. Dendritic cell subsets and lineages, and their functions in innate and adaptive immunity. Cell. 2001 Aug 10;106(3):259-62.
64. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014 Apr 18;344(6181):310-3.
65. Liu J, Zhang X, Chen K, Cheng Y, Liu S, Xia M, et al. CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity. 2019 Mar 19;50(3):600-15.
66. Ji J, Yin Y, Ju H, Xu X, Liu W, Fu Q, Hu J, Zhang X, Sun B. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death & Disease. 2018 Apr 30;9(5):478.
67. Cao J, Dong R, Jiang L, Gong Y, Yuan M, You J, et al. LncRNA-MM2P identified as a modulator of macrophage M2 PolarizationlncRNA-MM2P regulates M2 macrophage polarization. Cancer Immunology Research. 2019 Feb 1;7(2):292-305.
68. Ye Y, Xu Y, Lai YU, He W, Li Y, Wang R, Luo X, Chen R, Chen T. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. Journal of Cellular Biochemistry. 2018 Mar;119(3):2951-63.
69. Zhou YX, Zhao W, Mao LW, Wang YL, Xia LQ, Cao M, Shen J, Chen J. Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. The International Journal of Biochemistry & cell biology. 2018 Nov 1;104:25-33.
70. Zhou L, Tian Y, Guo F, Yu B, Li J, Xu H, et al. LincRNA-p21 knockdown reversed tumor-associated macrophages function by promoting MDM2 to antagonize* p53 activation and alleviate breast cancer development. Cancer Immunology, Immunotherapy. 2020 May;69:835-46.
71. Li X, Lei Y, Wu M, Li N. Regulation of macrophage activation and polarization by HCC-derived exosomal lncRNA TUC339. International Journal of Molecular Sciences. 2018 Sep 28;19(10):2958.
72. Xie C, Guo Y, Lou S. LncRNA ANCR promotes invasion and migration of gastric cancer by regulating FoxO1 expression to inhibit macrophage M1 polarization. Digestive Diseases and Sciences. 2020 Oct;65:2863-72.
73. Wu Y, Yi M, Niu M, Mei Q, Wu K. Myeloid-derived suppressor cells: an emerging target for anticancer immunotherapy. Molecular Cancer. 2022 Sep 26;21(1):184.
74. Zheng Y, Tian X, Wang T, Xia X, Cao F, Tian J, et al. Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Molecular Cancer. 2019 Dec;18:61.
75. Gao Y, Sun W, Shang W, Li Y, Zhang D, Wang T, et al. Lnc-C/EBPβ negatively regulates the suppressive function of myeloid-derived suppressor cells. Cancer Immunology Research. 2018 Nov;6(11):1352-63.
76. Gao Y, Wang T, Li Y, Zhang Y, Yang R. Lnc-chop promotes immunosuppressive function of myeloid-derived suppressor cells in tumor and inflammatory environments. The Journal of Immunology. 2018 Apr 15;200(8):2603-14.
77. Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nature Cell Biology. 2019 Apr;21(4):498-510.
78. Mi X, Xu R, Hong S, Xu T, Zhang W, Liu M. M2 macrophage-derived exosomal lncRNA AFAP1-AS1 and MicroRNA-26a affect cell migration and metastasis in esophageal cancer. Molecular Therapy-Nucleic Acids. 2020 Dec 4;22:779-90.
79. Chen Y, McAndrews KM, Kalluri R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nature Reviews Clinical Oncology. 2021 Dec;18(12):792-804.
80. Ding L, Ren J, Zhang D, Li Y, Huang X, Hu Q, et al. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis. 2018 Mar 8;39(3):397-406.
81. Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8(14):3932-48.
82. Deng X, Ruan H, Zhang X, Xu X, Zhu Y, Peng H, et al. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. International Journal of Cancer. 2020 Mar 15;146(6):1700-16.
83. Jin N, Jin N, Bu W, Li X, Liu L, Wang Z, et al. Long non-coding RNA TIRY promotes tumor metastasis by enhancing epithelial-to-mesenchymal transition in oral cancer. Experimental Biology and Medicine. 2020 Apr;245(7):585-96.
84. Liu T, Han C, Fang P, Ma Z, Wang X, Chen H, et al. Cancer-associated fibroblast-specific lncRNA LINC01614 enhances glutamine uptake in lung adenocarcinoma. Journal of Hematology & Oncology. 2022 Dec;15(1):141.
85. Li Y, Zhao Z, Liu W, Li X. SNHG3 functions as miRNA sponge to promote breast cancer cells growth through the metabolic reprogramming. Applied Biochemistry and Biotechnology. 2020 Jul;191(3):1084-99.
86. Leisegang MS, Fork C, Josipovic I, Richter FM, Preussner J, Hu J, et al. Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation. 2017 Jul 4;136(1):65-79.
87. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circulation Research. 2015 Mar 27;116(7):1143-56.
88. Parasramka MA, Maji S, Matsuda A, Yan IK, Patel T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacology & Therapeutics. 2016 May 1;161:67-78.
89. Gutschner T, Baas M, Diederichs S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011;21(11):1944-54.
90. Wheeler TM, Leger AJ, Pandey SK, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488(7409):111-5.
91. Hu Q, Ye Y, Chan LC, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol. 2019;20(7):835-51.
92. Mou X, Liew SW, Kwok CK. Identification and targeting of G-quadruplex structures in MALAT1 long non-coding RNA. Nucleic acids research. 2022;50(1):397-410.
93. Andergassen D, Rinn JL. From genotype to phenotype: genetics of mammalian long non-coding RNAs in vivo. Nature Reviews Genetics. 2022 Apr;23(4):229-43.
94. Zhang Y, Liu Q, Liao Q. Long noncoding RNA: a dazzling dancer in tumor immune microenvironment. J Exp Clin Cancer Res. 2020;39(1):231.
95. Winkle M, El-Daly SM, Fabbri M, et al. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20(8):629-51.
96. Pathania AS, Prathipati P, Challagundla KB. New insights into exosome mediated tumor-immune escape: Clinical perspectives and therapeutic strategies. Biochim Biophys Acta Rev Cancer. 2021;1876(2):188624.
97. He SW, Xu C, Li YQ, Li YQ, Zhao Y, Zhang PP, et al. AR-induced long non-coding RNA LINC01503 facilitates proliferation and metastasis via the SFPQ-FOSL1 axis in nasopharyngeal carcinoma. Oncogene. 2020 Aug 20;39(34):5616-32.
98. Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW, et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Research. 2019 Sep 15;79(18):4612-26.
99. Li ZX, Zheng ZQ, Yang PY, Lin L, Zhou GQ, Lv JW, et al. WTAP-mediated m6A modification of lncRNA DIAPH1-AS1 enhances its stability to facilitate nasopharyngeal carcinoma growth and metastasis. Cell Death & Differentiation. 2022 Jun;29(6):1137-51.
100. Wen X, Tang X, Li Y, Ren X, He Q, Yang X, et al. Microarray expression profiling of long non-coding RNAs involved in nasopharyngeal carcinoma metastasis. International Journal of Molecular Sciences. 2016 Nov 23;17(11):1956.
101. Liang YL, Zhang Y, Tan XR, Qiao H, Liu SR, Tang LL, et al. A lncRNA signature associated with tumor immune heterogeneity predicts distant metastasis in locoregionally advanced nasopharyngeal carcinoma. Nature Communications. 2022 May 30;13(1):2996.
102. Li Y, Jiang T, Zhou W, Li J, Li X, Wang Q, et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nature Communications. 2020 Feb 21;11(1):1000.
103. Zhang Y, Zhang L, Xu Y, Wu X, Zhou Y, Mo J. Immune-related long noncoding RNA signature for predicting survival and immune checkpoint blockade in hepatocellular carcinoma. Journal of Cellular Physiology. 2020 Dec;235(12):9304-16.
104. Wang J, Shen C, Dong D, Zhong X, Wang Y, Yang X. Identification and verification of an immune-related lncRNA signature for predicting the prognosis of patients with bladder cancer. International Immunopharmacology. 2021 Jan 1;90:107146.
105. Sun J, Zhang Z, Bao S, Yan C, Hou P, Wu N, et al. Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. Journal for Immunotherapy of Cancer. 2020;8(1).
106. Zhou M, Zhang Z, Bao S, Hou P, Yan C, Su J, et al. Computational recognition of lncRNA signature of tumor-infiltrating B lymphocytes with potential implications in prognosis and immunotherapy of bladder cancer. Briefings in Bioinformatics. 2021 May;22(3):bbaa047.
107. Liu Z, Liu L, Weng S, Guo C, Dang Q, Xu H, et al. Machine learning-based integration develops an immune-derived lncRNA signature for improving outcomes in colorectal cancer. Nature Communications. 2022 Feb 10;13(1):816.
108. Wang L, Felts SJ, Van Keulen VP, Scheid AD, Block MS, Markovic SN, et al. Integrative genome-wide analysis of long noncoding RNAs in diverse immune cell types of melanoma PatientsCharacterization of long noncoding RNAs in melanoma. Cancer Research. 2018 Aug 1;78(15):4411-23.
109. Zhou J, Xu J, Zhang L, Liu S, Ma Y, Wen X, et al. Combined single-cell profiling of lncRNAs and functional screening reveals that H19 is pivotal for embryonic hematopoietic stem cell development. Cell Stem Cell. 2019 Feb 7;24(2):285-98.e5.
110. Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biology. 2016 Dec;17:67.
111. Vallot C, Patrat C, Collier AJ, Huret C, Casanova M, Ali TM, et al. XACT noncoding RNA competes with XIST in the control of X chromosome activity during human early development. Cell Stem Cell. 2017 Jan 5;20(1):102-11.
112. Aznaourova M, Schmerer N, Janga H, Zhang Z, Pauck K, Bushe J, et al. Single-cell RNA sequencing uncovers the nuclear decoy lincRNA PIRAT as a regulator of systemic monocyte immunity during COVID-19. Proceedings of the National Academy of Sciences. 2022 Sep 6;119(36):e2120680119.
113. Wang Y, Huang L, Wang Y, Luo W, Li F, Xiao J, et al. Single-cell RNA-sequencing analysis identifies host long noncoding RNA MAMDC2-AS1 as a co-factor for HSV-1 nuclear transport. International Journal of Biological Sciences. 2020;16(9):1586-603.
114. Singh AV, Kayal A, Malik A, Maharjan RS, Dietrich P, Thissen A, et al. Interfacial Water in the SARS Spike Protein: Investigating the Interaction with Human ACE2 Receptor and In Vitro Uptake in A549 Cells. Langmuir. 2022 Jun 23;38(26):7976-88.
115. Chandrasekar V, Singh AV, Maharjan RS, Dakua SP, Balakrishnan S, Dash S, et al. Perspectives on the Technological Aspects and Biomedical Applications of Virus-Like Particles/Nanoparticles in Reproductive Biology: Insights on the Medicinal and Toxicological Outlook. Advanced NanoBiomed Research. 2022 Aug;2(8):2200010.
116. Li X, Meng X, Wei C, Zhou Y, Chen H, Huang H, Chen M. Dissecting LncRNA Roles in Renal Cell Carcinoma Metastasis and Characterizing Genomic Heterogeneity by Single-Cell RNA-seqLncRNA Profiling of RCC Metastasis. Molecular Cancer Research. 2018 Dec 1;16(12):1879-88.
117. Xu M, Zhou C, Weng J, Chen Z, Zhou Q, Gao J, et al. Tumor associated macrophages-derived exosomes facilitate hepatocellular carcinoma malignance by transferring lncMMPA to tumor cells and activating glycolysis pathway. Journal of Experimental & Clinical Cancer Research. 2022 Aug 19;41(1):253.
118. Dlamini Z. Artificial Intelligence and Precision Oncology: Bridging Cancer Research and Clinical Decision Support. Springer Cham. 2023, https://doi.org/10.1007/978-3-031-21506-3.
119. Tibshirani R. The lasso method for variable selection in the Cox model. Statistics in Medicine. 1997 Feb 28;16(4):385-95.
120. Koelzer VH, Sirinukunwattana K, Rittscher J, Mertz KD. Precision immunoprofiling by image analysis and artificial intelligence. Virchows Archiv. 2019 Apr 1;474:511-22.
121. Dakua SP, Abi-Nahed J. Patient oriented graph-based image segmentation. Biomedical Signal Processing and Control. 2013 May 1;8(3):325-32.
122. Dakua SP, Abinahed J, Al-Ansari AA. Pathological liver segmentation using stochastic resonance and cellular automata. Journal of Visual Communication and Image Representation. 2016 Jan 1;34:89-102.
123. Dakua SP. Use of chaos concept in medical image segmentation. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization. 2013 Mar 1;1(1):28-36.
124. Horai Y, Mizukawa M, Nishina H, Nishikawa S, Ono Y, Takemoto K, et al. Quantification of histopathological findings using a novel image analysis platform. Journal of Toxicologic Pathology. 2019;32(4):319-27.
125. Singh AV, Maharjan RS, Kromer C, Laux P, Luch A, Vats T, et al. Advances in smoking related in vitro inhalation toxicology: A perspective case of challenges and opportunities from progresses in lung-on-chip technologies. Chemical Research in Toxicology. 2021 Aug 16;34(9):1984-2002.
126. Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L, et al.Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014 Dec;14(1):693.
127. Zhang M, Zheng Y, Sun Y, Li S, Chen L, Jin X, et al. Knockdown of NEAT1 induces tolerogenic phenotype in dendritic cells by inhibiting activation of NLRP3 inflammasome. Theranostics. 2019;9(12):3425-42.
128. Huan L, Guo T, Wu Y, Xu L, Huang S, Xu Y, et al. Hypoxia induced LUCAT1/PTBP1 axis modulates cancer cell viability and chemotherapy response. Molecular Cancer. 2020 Dec;19:11.
129. Zheng A, Song X, Zhang L, Zhao L, Mao X, Wei M, et al. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/β-catenin pathway. Journal of Experimental & Clinical Cancer Research. 2019 Dec;38(1):305.
130. Agarwal S, Vierbuchen T, Ghosh S, Chan J, Jiang Z, Kandasamy RK, et al. The long non-coding RNA LUCAT1 is a negative feedback regulator of interferon responses in humans. Nature Communications. 2020 Dec 11;11(1):6348.
131. Yuan X, Shen Q, Ma W. Long Noncoding RNA hotair promotes the progression and immune escape in laryngeal squamous cell carcinoma through MicroRNA-30a/GRP78/PD-L1 Axis. Journal of Immunology Research. 2022 Apr 4;2022: 5141426.
132. Wang Y, Yi K, Liu X, Tan Y, Jin W, Li Y, et al. HOTAIR up-regulation activates NF-κB to induce immunoescape in gliomas. Frontiers in Immunology. 2021 Nov 23;12:785463.
133. Obaid M, Udden SM, Deb P, Shihabeddin N, Zaki M, Mandal SS. LncRNA HOTAIR regulates lipopolysaccharide-induced cytokine expression and inflammatory response in macrophages. Scientific Reports. 2018 Oct 23;8(1):15670.
134. Obaid M, Udden SN, Alluri P, Mandal SS. LncRNA HOTAIR regulates glucose transporter Glut1 expression and glucose uptake in macrophages during inflammation. Scientific Reports. 2021 Jan 8;11(1):232.
135. Yao Z, Xiong Z, Li R, Liang H, Jia C, Deng M. Long non-coding RNA NRON is downregulated in HCC and suppresses tumour cell proliferation and metastasis. Biomedicine & Pharmacotherapy. 2018 Aug 1;104:102-9.
136. Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005 Sep 2;309(5740):1570-3.
