Abstract
Dysfunction and abnormal differentiation of immune cells and hematopoietic precursors are well described in patients with cancer, although the role of transcription factors in these defects is not well established. Here, we evaluated expression of C/EBPa, PU.1 and BACH1 transcription factors in lymphoid and myeloid cells, including NK cells, T cells, neutrophils and monocytes, isolated from healthy donors and cancer patients. The results revealed a significant suppression of transcription factors participating in the development of immune cells in patients with cancer. Analysis of expression of transcription factors in CD34+CD45dim hematopoietic precursors showed that in contrast to the downregulation of transcription factor in mature peripheral blood immune cells, expression of these regulators in stem/precursor cells was significantly increased in patients with lung cancer, especially in patients with detectable metastases. These results open a new opportunity to test whether altered transcriptional regulation of mature immune cells in cancer may be intrinsically related to abnormal functioning of transcription factors in early hematopoiesis or occurs independently via direct effects of tumor-derived factors on circulating immune cells.
Keywords
NK cells, T cells, neutrophils, monocytes, C/EBPα, BACH1, PU.1
Introduction
Cancer remains one of the leading causes of death worldwide despite a significant progress in cancer diagnostics, treatment and prophylactics during last decades. It is generally accepted that the development and progression of cancer are closely associated with dysregulated functioning and diminishing of local and systemic immune responsiveness. Immune abnormalities in cancer may be due to the effect of tumor/stroma-derived factors on mature circulating immune cells or on their precursors in the bone marrow. The hypothesis of intrinsic transcriptional defects in the differentiation mechanism of hematopoietic stem and progenitor cells as a source of dysfunction of immature and mature immune cells in cancer patients is supported by a growing number of recent findings. A significant reduction in c-myc expression was observed in both NK cells and T lymphocytes in patients with cancer [1,2]. There is evidence of a high level of c-myc expression in neutrophils in cancer [3]. Cancer-associated abnormalities in differentiation of hematopoietic progenitor cells and in myeloid and lymphoid cells have also been reported [4-7]. Thus, it is conceivable to speculate that an abnormal expression of proto-oncogenes in lymphoid cells and myeloid cells may be due to a general imbalance of transcription factors responsible for the lineage-specific differentiation.
Leukocytes’ fate is progressively determined during the gradual differentiation of hematopoietic stem cells (HSCs) into multipotent progenitors (MPPs) and common lymphoid or myeloid progenitors [8]. Transcription factors are key regulators of these cells acting through their capacity to bind DNA and control gene transcription, which thus regulates the development of specific hematopoietic lineages from stem cells. Among transcription factors C/EBPα/β, BACH1/2 and ETS play pivotal roles in myeloid and lymphoid cell differentiation. However, the gradual mechanisms by which differentiation of common progenitors controlled under cancerous condition have been unclear.
C/EBPΑ (CCAAT/enhancer binding protein (C/EBP) alpha), a protein encoded by the C/EBPΑ gene in humans, was originally identified 30 years ago as a transcription factor that binds both promoter and enhancer regions. Analysis of its function in hematopoietic malignancies as well as in solid cancer has revealed many ways how cancerous cells abrogate C/EBPα function [9-11]. Although early studies focused on the role of C/EBPα in regulating transcriptional and differentiation processes during liver and adipogenic development, recent reports provide more mechanistic insights into its role in normal and malignant hematopoiesis [12,13]. For instance, C/EBPα has been reported to protect adult HSCs from apoptosis and to maintain their quiescent state. Consequently, deletion of C/EBPΑ is associated with loss of self-renewal and HSC exhaustion [14] as well as the absence of mature neutrophils and eosinophils in mutant mice [15]. Thus, C/ EBPα functions as a priming factor at the HSC level where it supports myeloid differentiation and reduces lymphoid lineage selection, which forms the epigenetic shape of HSCs for balancing various cell fate choices.
Analysis of C/EBPα target genes essential for myeloid cell development identified c-myc as a C/EBPα negatively regulated gene in myeloid cell lines [16]. The fact that C/EBPα directly affects the level of c-myc expression and, thus, the granulocytic differentiation pathway, is intriguing as it has been previously shown that downregulation of c-myc can induce myeloid differentiation [17]. As proliferation and differentiation are mutually exclusive, c-myc - a proliferative factor, and C/EBPα - a differentiation factor, work in opposition to each other. Given that c-myc expression is down-regulated in NK and T cells in patients with cancer [1,2,18], this raises an interesting question about C/EBPα activity in different lymphocyte subsets in cancer.
PU.1 (Spi1, E26-transformation-specific (ETS) factor) is another transcription factor requiring for the development of the immune system that functions at both early and late stages of lymphoid and myeloid differentiation [19]. The Pu.1 gene specifically expressed in the early stages of hematopoiesis and its knockout generates mice that lack both myeloid and lymphoid cells [20]. PU.1 mRNA and protein expression is detectable in HSCs, mature monocytes, granulocytes, B, NK cells and pre-T cells [21-23]. Low-level expression of PU.1 in hematopoietic precursors induces B cell differentiation, whereas high levels favor myeloid differentiation [24]. Interestingly, the differentiation potential of T lineage cells becomes restricted soon after entry of multipotent precursors into the thymus and is accompanied by a downregulation of C/ EBPα and PU.1 [25].
Furthermore, C/EBPα cooperates with PU.1 to regulate myeloid gene expression [26]. For instance, in pre-B cells, C/EBPα binds to both pre-existing enhancers occupied by PU.1 and de novo enhancers, and the combined activation of these enhancers recapitulates the activation of myeloid enhancers and associated genes during normal hematopoiesis [27]. Similar results were obtained in experiments with inducible expression of C/EBPα and PU.1 in pre-T cells: PU.1 converted them into myeloid dendritic cells, while C/EBPα induced the formation of macrophages, which required endogenous PU.1 [25]. Moreover, Notch signaling induced T lineage restriction by downregulating C/EBPα and PU.1 in multilineage precursors [25]. These results together with the findings of altered Notch 1 and Notch 2 expression in NK cells in patients with cancer [2,18] justify further analysis of C/ EBPα and PU.1 transcription factors in lymphocytes and hematopoietic precursors isolated from cancer patients.
BACH1 and BACH2 (BTB and CNC homology) are transcription repressors that belong to the basic regionleucine zipper family [28]. With the ability to bind to heme, BACH1 and BACH2 are important in maintaining heme homeostasis in response to oxidative stress [29]. BACH1 and BACH2 promote the differentiation of common lymphoid progenitors to B cells by repressing myeloidrelated genes [30]. BACH1-/-BACH2-/- double knockout mice show erythropoiesis disorders with increased myelopoiesis from common myeloid progenitors [31]. BACH2 has critical roles in both acquired immunity and innate immunity, including early B cell development and immunoglobulin class-switch recombination, the development of effector and regulatory T cells and the activation of tissue macrophages [32-35].
BACH1 and BACH2 have important, non-redundant functions in mature myeloid and lymphoid lineages, which are due to partial mutual exclusivity in their patterns of expression: BACH2 mRNA is mostly expressed in mature lymphoid lineages, whereas BACH1 mRNA is generally expressed in myeloid lineages [36]. Intriguingly, BACH2 represses C/EBPβ and C/EBPβ represses BACH2, and both of them bind to overlapping regulatory regions at their myeloid target genes, suggesting the presence of a mutual feedforward loop leading to myeloid gene regulation [37]. Although, the involvement of BACH2-C/EBPβ pathway in the HSC commitment to myeloid and lymphoid lineages at normal conditions and after infection has thus been shown, little is known about changes of BACH and C/EBP transcription factors in peripheral blood hematopoietic precursors and leukocytes in patients with different types of solid tumors.
The studies described in this report continue to investigate the state of transcription factor expression in immune cells and their precursors in patients with cancer. Our results revealed a significant down-regulation of PU.1 in NK cells and BACH1 in T cells in all cancer patients. However, C/ EBPα decrease was detected in monocytes in patients with gastric cancer and neutrophils in patients with lung cancer. Unexpectedly, we revealed a significant increase in the expression of C/EBPα, PU.1 and BACH1 transcription factors in HSC in patients with lung but not gastric cancer. These results demonstrate that abnormal transcriptional regulation of hematopoietic stem cells and differentiated immune cells is cell and cancer dependent and provide new insights into the potential roles of different immune transcriptional factors in tumor escape mechanisms. Although these results are promising, further investigation is justified to identify possible factors responsible for altered expression of transcriptional factors in immune and stem cells in patients with cancer.
Results
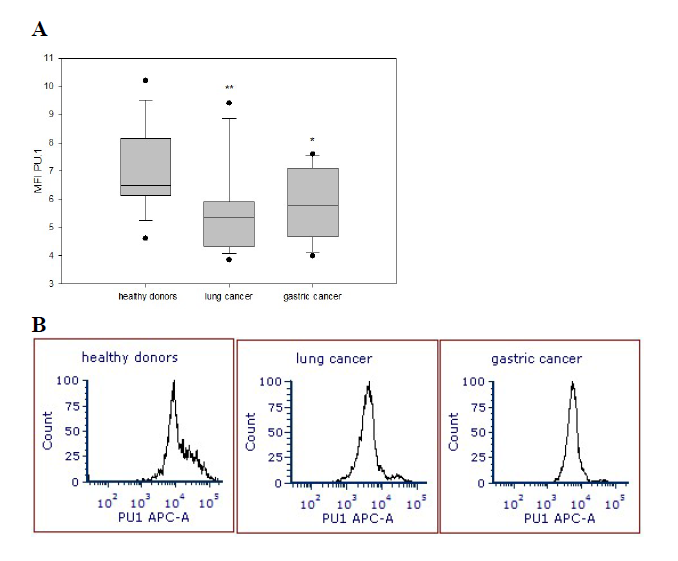
Decrease in the PU.1 expression in NK cells in cancer patients
Intracellular expression of C/EBPα, PU.1 and BACH1 transcription factors was determined in the peripheral blood NK cells isolated from patients with lung and gastric cancer and healthy donors utilizing specific antibodies and flow cytometry. MFI data revealed that the expression of PU.1 transcription factor in NK cells in patients with lung cancer (5.55 ± 0.42) and gastric cancer (5.81 ± 0.32) was significantly reduced compared with the expression of PU.1 in NK cells harvested from healthy volunteers (6.99 ± 0.37) (p<0.003 and p<0.02, respectively) (Figure 1). At the same time, expression of C/EBPα and BACH1 transcription factors in NK cells in cancer patients was not different from their expression in NK cells from healthy donors.
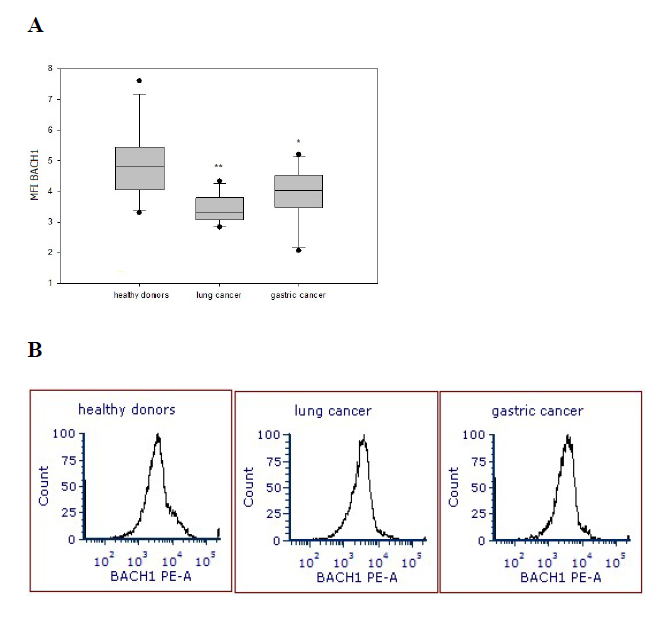
Reduced BACH1 expression in T cells in cancer patients
Expression of C/EBPα, PU.1 and BACH1 transcription factors also was determined in the peripheral blood T cells isolated from patients with lung cancer and gastric cancer and healthy donors. In contrast to NK cell results, in T cells, only expression of BACH1 was significantly diminished in all patients regardless the type of cancer, tumor cell differentiation stage, clinical stage of the disease or the presence of metastases. MFI data revealed that the expression of BACH1 transcription factor in T cells in patients with lung cancer (3.43 ± 0.13) and gastric cancer (3.99 ± 0.24) was also significantly reduced compared with healthy volunteers (4.98 ± 0.31) (p<0.001 and p<0.02, respectively) (Figure 2).
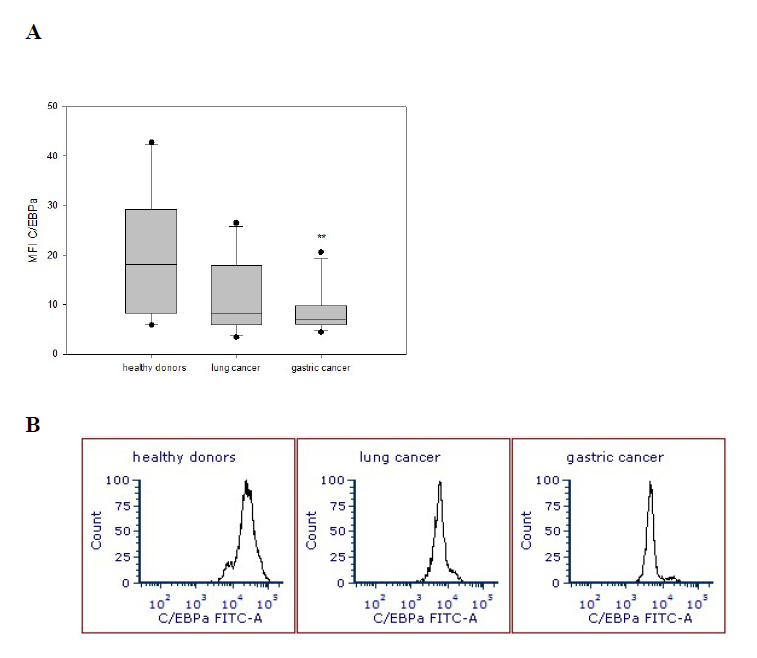
Differential expression of transcription factors in monocytes in patients with different cancer localization
The expression of transcription factors in the peripheral blood monocytes was also analyzed. C/EBPα transcription factor expression in these cells was significantly reduced in patients with gastric cancer (8.77 ± 1.27) but not lung cancer compared with the expression seen in healthy volunteers (20.23 ± 3.51) (p<0.006) (Figure 3). With respect to transcription factors PU.1 and BACH1, their expression was not significantly reduced in any cancer patient tested cohorts when compared with the results obtained from healthy donors.
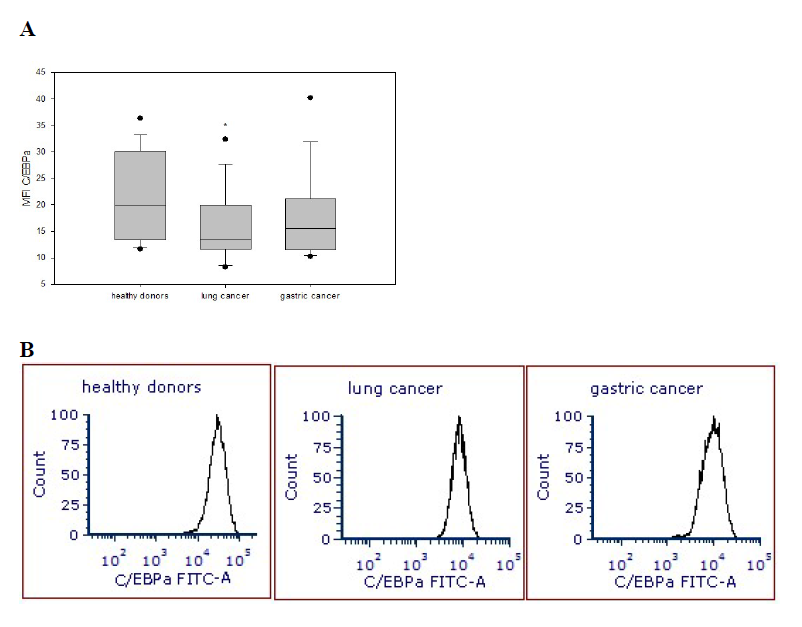
Expression of transcription factors in neutrophils
Next, analyzing expression of transcription factors in neutrophils, we revealed a common trend of their downregulation in cancer in comparison to healthy cells. However, this decrease reached a significant level only for C/EBPα expression in neutrophils obtained from patients with lung cancer (15.48 ± 1.74) compared with healthy volunteers (21.43 ± 2.05) (p=0.043) (Figure 4). The expression of BACH1 and PU.1 transcription factors in neutrophils in cancer patients was not significantly altered when compared with healthy donors.
Up-regulation of the expression of transcription factors in CD34+CD45dim cells
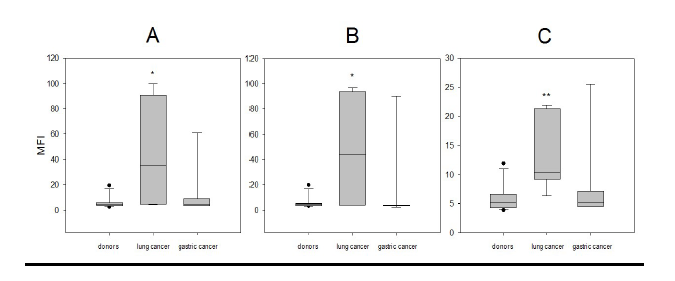
CD34 is a transmembrane glycoprotein expressed on early lymphohematopoietic stem and progenitor cells and CD34+CD45dim cells are early stem/progenitor cells [38,39]. Analysis of these cells in the peripheral blood of cancer patients and healthy donors demonstrated a significant increase in the expression of all three transcription factors in patients with lung cancer. For instance, the MFI levels of C/EBPα (44.69 ± 18.35), BACH1 (47.64 ± 19.41) and PU.1 (13.37 ± 2.65) in stem/progenitor cells from patients with lung cancer were significantly higher than in healthy volunteers: C/EBPα (5.63 ± 1.43), BACH1 (5.59 ± 1.44) and PU.1 (5.89 ± 0.68) (p=0.021, p=0.044 and p<0.003, respectively) (Figure 5). No significant differences were detected in transcriptional factor expression in stem/ progenitor cells from patients with gastric cancer.
Discussion
Lineage-instructive transcription factors trigger and subdue cell-specific genes by identifying sequence-specific DNA consensus motifs enclosed within enhancers and promoters [40]. Though, how these processes are matched in early and late myeloid and lymphoid differentiation in the tumor environment remain unclear especially as neither transcription factor-induced lineage conversions nor MPP/HPC reprogramming seem to repeat normal differentiation paths during carcinogenesis. Whereas lymphoid and myeloid effector cells possess undisputable tumor-suppressive properties across multiple types of cancer, it is well known that their antitumor properties and function are compromised in cancer patients [41-44]. Established dependence of immune cell development on given transcription factors allows speculating that in the tumor or even pre-tumor environment abnormal functioning of immune effectors is the result of an aberrant differentiation of hematopoietic precursors due to a compromised regulation of transcription factors.
Our previous results revealed multiple defects in c-kit/ SCF signaling and c-Myc expression in NK and T cells as well as reduced Notch signaling in NK cells in cancer patients [1,45]. This suggests that a decrease in protooncogene expression may be a consequence of a defect in transcription factors associated with their regulation. The c-kit and its ligand SCF play an important role in survival and cytotoxicity of NK cells and differentiation of NK cells from the precursors is reduced in the absence of c-kit signaling [46]. Furthermore, PU.1 plays an intrinsic role in NK cell commitment from lymphoid progenitor, regulates cell development and production of IFN-γ [47], and a significant down-regulation of the c-kit expression in PU.1 deficient NK cells has been reported [48]. In addition, the fraction of cycling NK cells is significantly decreased in PU.1-/- chimeras [48]. We have also described a mitotic arrest of NK cells in G0/G1 phase of the cell cycle and decreased amount of NK cells in phases G2/M and S in cells isolated from patients with lung and gastric cancer [1]. Our new results here revealed a significant reduction in PU.1 expression in NK cells from patients with cancer, which correlates with the previously reported down-regulation of c-kit expression and cell mitotic arrest in the same cells. Therefore, it is possible that previously described molecular abnormalities in NK cells in cancer reflect a single regulatory or developmental pathway that is primarily disrupted in cancer patients.
Our results here demonstrated similar alterations of other transcription factors in isolated lymphocytes. For instance, expression of BACH1, playing an important role in T cell development [37,49], was significantly lower in T cells in cancer patients. Together with the indications of T cell exhaustion in cancer patients [50] and reported downregulation of c-myc expression in T cells from patients with cancer, which can lead to metabolic defects in T cells [1,51], our new results support the notion about a potential transcriptional dysregulation in lymphoid cells and their precursor cells during carcinogenesis. Further, a growing number of evidences demonstrating that an increased peripheral neutrophil to lymphocyte ratio significantly associated with poorer outcome in various cancers [52-54], our results raised a question about transcription factor expression in myeloid cells in cancer.
The network governing neutrophil-macrophage progenitor cell fate has several key determinants, such as myeloid master regulators C/EBPα and PU.1. Here, we observed decreased expression of granulo-monocytic transcription factor C/EBPα in both monocytes and neutrophils isolated from patients with cancer, while no significant alterations of expression of BACH1 and PU.1 were determined. Interestingly, a decrease in C/EBPα expression in monocytes was observed only in patients with gastric cancer while in neutrophils it was only seen in patients with lung cancer. Although there is no clear explanation for this phenomenon, one can suggest that C/EBPα expression is affected on the level of neutrophilmonocyte progenitor phenotype. However, this ectopic neutrophil-monocyte potential can be transcriptionally or epigenetically reprogrammed in a cancer-specific manner defining the lineage-specific identity of the resulting myeloid malformation. Interestingly, down-regulation of C/EBPα may be induced epigenetically by a lysine-specific demethylase-1 (LSD1) known to be crucial for human leukemogenesis and currently being one of the emerging therapeutic targets [55]. Pharmacological inhibition of factors which control balancing of lineage-specific transcription factors, like LSD1, for instance, should cause lineage-dependent metabolic remodeling in immune cell differentiation. This should open new opportunity in harnessing development of myeloid effector and regulatory cells in the tumor milieu.
Nonetheless, more regulators remain to be identified and characterized under the tumor growth conditions. Another possibility is to characterize transcriptional regulation in the early progenitor cells isolated from cancer patients. Here, we have tested the CD34+CD45dim stem//progenitor cells in the peripheral blood and obtained unexpected results. The up-regulated expression of all tested transcription factors in CD34+CD45dim cells was identified in patients with lung cancer. A particularly high increase was observed in four patients with combined lung and colon or breast cancers and metastases (data not shown). These intriguing results require further verification in bigger cohorts of patients with different types of cancer.
Although C/EBPα plays a well-characterized role in the formation of myeloid progenitors [14] and elicits a lineageinstructive function in MPPs towards the myeloid lineage [12], BACH1/2 regulate both commitment and on-demand hematopoiesis because they control erythroid-myeloid and lymphoid-myeloid differentiation by repressing the myeloid program [56], and PU.1 displays indispensable and distinct roles in hematopoietic development through supporting HSC self-renewal and commitment and maturation of myeloid and lymphoid lineages [57], very little is known about the biological and clinical significance of these transcriptional factor natural up-regulation in hematopoietic progenitor/stem cells especially during carcinogenesis. TNF-α is the principal PU.1 inducing signal in HSCs: it can directly and rapidly upregulate PU.1 protein in HSCs in vitro and in vivo and is both sufficient and required to relay signals from inflammatory challenges to HSCs [58]. M-CSF (also called CSF1), a myeloid cytokine released during inflammation, can also directly induce activation of the PU.1 promoter and instruct myeloid cellfate change in HSCs in vitro and in vivo [59]. Although genetic changes in transcription factor balance are known to sensitize HSCs to cytokine instruction, the initiation of HSC commitment is generally thought to be prompted by stochastic variation in cell-intrinsic regulators such as lineage-specific transcription factors, leaving cytokines to ensure survival and proliferation of the progeny cells. However, the abovementioned facts demonstrate that HSCs are susceptible to direct lineage-specifying effects of cytokines. Therefore, it is possible that tumor/ stroma-derived factors may directly alter transcription factors in the early hematopoietic progenitors, as we detected in our studies.
In summary, our results demonstrate altered expression of different transcription factors in lymphoid and myeloid cells in patients with different types of cancer. These changes may be associated with internal dysregulation of hematopoietic stem/progenitor cells induced by early or progressing tumors. The origin and link between these phenomena remain to be determined.
Materials and Methods
Patients’ samples
Peripheral blood samples were collected from patients with lung and GI cancers (Table 1). Blood specimens were collected prior to all surgical or chemotherapy procedures. Healthy controls (n = 23), nine men and fourteen women with median age 54 (range 42–68) were recruited from the personnel of the laboratories of the Research Institute of Fundamental and Applied Medicine. All studies were approved by the Institutional Review Board and all patients and healthy donors provided signed written informed consent for sample acquisition for research purposes.
| Characteristics | |
| Total number | 43 |
| Gender, N (%) | |
| Male | 25 (58%) |
| Female | 18 (42%) |
| Age, years | |
| Median | 62 |
| Range | 23-82 |
| Prior anticancer therapies | 0 |
| Lung cancer | 20 (46%) |
| NSCLC | 17 (85%) |
| Adenocarcinoma | 15 (75%) |
| Squamous cell carcinoma | 2 (10%) |
| GI cancer | 23 (54%) |
| Esophageal ADC | 6(26%) |
| Colorectal cancer | 6 (26%) |
| Sigmoid cancer | 2 (8.6%) |
| Gastric ADC | 9 (39.1%) |
| Metastasis | |
| Primary tumor site | Localization of metastasis |
| Lung ADC (4) | Bones, colon, breast, medias- tinum |
| Colorectal cancer (3) | Lymph nodes |
Isolation of NK cells, CD3+ T cells and monocytes
PBMC were isolated from the peripheral blood by Ficoll- Paque PLUS (Life Technologies, USA) density gradient centrifugation. NK cells were negatively selected using DynaMag Magnet with Dynabeads Untouched Human NK Cells Isolation kit (Life Technologies). T cells were isolated from PBMC by positive selection using DynaMag Magnet with Dynabeads Untouched Human CD3 microbeads (Life Technologies). Monocytes were isolated from PBMC by positive selection using DynaMag Magnet with Dynabeads™ CD14 microbeads (Invitrogen). The purity of the cell subsets was confirmed by flow cytometry (FACSCanto II, BD Biosciences) using specific CD56 and CD16 monoclonal antibodies conjugated with FITC and PerCP, specific CD3 monoclonal antibodies labeled with FITC and specific CD14 monoclonal antibodies labeled with PE (BD Biosciences). Data analysis was performed with BD FACSDiva Software.
Isolation of neutrophils
Neutrophils were isolated using the method that allows fast isolation of untouched neutrophils without density gradient centrifugation. Erythrocytes in the peripheral blood samples were aggregated and sedimented and neutrophils were removed by immunomagnetic purification with MACSxpress Beads (MACSxpress® Whole Blood Human Neutrophil Isolation Kit, Miltenyi Biotec Inc.). Phenotype of neutrophils was confirmed with CD15 expression by flow cytometry. All experimental procedures were optimized using blood samples from healthy donors.
Gating strategy for cell analysis
Gating strategy for NK cell analysis: the first step is cutting off dead cells on the FSC, then, in the area of lymphocytes, NK cells were identified based on FSC/ SSC. The purity of isolated NK cells after isolation and enrichment procedures was in the range of 89-95%.
Gating strategy of CD3 positive cell analysis: the first step is cutting off dead cells on the FSC, second step - in the area of lymphocytes; T cells were identified based on FSC/SSC. The purity of isolated CD3 cells after isolation and enrichment procedures was in the range of 93-96%.
Gating strategy of monocyte cell analysis: the first step is cutting off dead cells on the FSC, then monocytes cells were identified based on FSC/SSC and CD14 expression in the area of monocytes. The purity of the isolated monocytes after isolation and enrichment procedures was within the range of 87-94%.
Gating strategy of neutrophils cell analysis: the first step is cutting off dead cells on the FSC, and then neutrophils were identified based on FSC/SSC and CD15 expression in the area of granulocytes. The purity of the isolated neutrophils after isolation and enrichment procedures was within the range of 90-93%.
Gating strategy of CD34CD45dim cell analysis: the first step is cutting off dead cells on the FSC, cells are gated in the CD45dim-lymphocyte area, then 7-aminoactinomycin D (7-AAD)-positive dead cells were separated from viable CD34 positive 7-AAD-negative cells.
Detection of C/EBPα, PU.1 and BACH1 protein expression
For the intracellular detection of C/EBPα, PU.1 and BACH1 transcription factor expression by flow cytometry, isolated NK cells, T cells, neutrophils and monocytes were fixed, permeabilized and stained with differentially conjugated mouse monoclonal anti-human C/EBPα, PU.1 and BACH1 antibodies: C/EBPα (D-5) Alexa Fluor® 488 sc-365318 AF488, 200 μg/ml (RUO), BACH1 (F-9) PE (RUO), PU.1 (C-3) Alexa Fluor® 488, sc-390405 AF488 200 μg/ml (RUO) (Santa Cruz Biotechnology, Inc.).
Expression of transcription factors in the stem/ progenitor cells
Expression of C/EBPα, PU.1 and BACH1 was determined in the peripheral blood cell samples gated on CD34+ CD45dim cells by flow cytometry using conjugated mouse monoclonal anti-human C/EBPα, PU.1 and BACH1 antibodies according to the manufacturer’s protocols. Cells were analyzed by a 10-color FACSCanto (Becton Dickinson). CD45 was used as a marker of blood hematopoietic cells to exclude mature erythrocytes and platelets. CD34+CD45dim cells were considered stem/progenitor cells [60,61]. CD34 PE-Cy7 and CD45 PerCP-Cy5 antibodies were used for cell phenotyping. Data analysis was performed with BD FACSDiva Software.
Statistical analysis
For a single comparison of two groups, the Student’s t-test was used after the evaluation of normality. If data distribution was not normal, a Mann-Whitney rank sum test was performed. For the comparison of multiple groups, analysis of variance (ANOVA) was applied. SigmaStat Software was used for data analysis (SyStat Software Inc.). For all statistical analyses, p<0.05 was considered significant. All experiments were repeated at least two times. Data are presented as the mean ± SEM.
Acknowledgements
We would like to thank Sergey Bernikov for his crucial help in formal data analysis, application of statistical, mathematical, computational and other techniques to analyze and compare the results of this study.
Author Contributions
Conceptualization, writing, original draft preparation, Gulnur K. Zakiryanova; methodology, project administration, supervision, Narymzhan N. Nakisbekov; formal analysis, investigation, data curation, Elena Kustova, Nataliya T. Urazalieva, Galina V. Shurin; resources, Emile T. Baimuchametov, Valeriy A. Makarov; writing, review and editing, visualization, funding acquisition, Michael R. Shurin.
Funding
This research was funded by the Committee on Science of the Ministry of Education and Science of the Republic of Kazakhstan.
Conflicts of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
2. Zakiryanova GK, Kustova E, Urazalieva NT, Baimukhametov ET, Makarov VA, Turaly GM, et al. Notch signaling defects in NK cells in patients with cancer. Cancer Immunology, Immunotherapy. 2021 Apr;70(4):981-8.
3. Jablonska J, Leschner S, Westphal K, Lienenklaus S, Weiss S. Neutrophils responsive to endogenous IFN-β regulate tumor angiogenesis and growth in a mouse tumor model. The Journal of Clinical Investigation. 2010 Apr 1;120(4):1151-64.
4. Richards DM, Hettinger J, Feuerer M. Monocytes and macrophages in cancer: development and functions. Cancer Microenvironment. 2013 Aug 1;6(2):179-91.
5. Richards JO, Chang X, Blaser BW, Caligiuri MA, Zheng P, Liu Y. Tumor growth impedes natural-killercell maturation in the bone marrow. Blood. 2006 Jul 1;108(1):246-52.
6. Zhu YP, Padgett L, Dinh HQ, Marcovecchio P, Blatchley A, Wu R, et al. Identification of an early unipotent neutrophil progenitor with pro-tumoral activity in mouse and human bone marrow. Cell Reports. 2018 Aug 28;24(9):2329-41.
7. Casbon AJ, Reynaud D, Park C, Khuc E, Gan DD, Schepers K, et al. Invasive breast cancer reprograms early myeloid differentiation in the bone marrow to generate immunosuppressive neutrophils. Proceedings of the National Academy of Sciences. 2015 Feb 10;112(6):E566- 75.
8. Adolfsson J, Månsson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of Flt3+ lymphomyeloid stem cells lacking erythro-megakaryocytic potential: a revised road map for adult blood lineage commitment. Cell. 2005 Apr 22;121(2):295-306.
9. Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPα differentiation pathway in human cancer. Journal of Clinical Oncology. 2009 Feb 1;27(4):619.
10. Lourenço AR, Coffer PJ. A tumor suppressor role for C/EBPα in solid tumors: more than fat and blood. Oncogene. 2017 Sep;36(37):5221-30.
11. Schuster MB, Porse BT. C/EBPα: A tumour suppressor in multiple tissues?. Biochimica et Biophysica Acta (BBA)- Reviews on Cancer. 2006 Aug 1;1766(1):88-103.
12. Wölfler A, Danen-van Oorschot AA, Haanstra JR, Valkhof M, Bodner C, Vroegindeweij E, et al. Lineage-instructive function of C/EBPα in multipotent hematopoietic cells and early thymic progenitors. Blood, The Journal of the American Society of Hematology. 2010 Nov 18;116(20):4116-25.
13. Ohlsson E, Schuster MB, Hasemann M, Porse BT. The multifaceted functions of C/EBPα in normal and malignant haematopoiesis. Leukemia. 2016 Apr;30(4):767-75.
14. Hasemann MS, Lauridsen FK, Waage J, Jakobsen JS, Frank AK, Schuster MB, et al. C/EBPα is required for longterm self-renewal and lineage priming of hematopoietic stem cells and for the maintenance of epigenetic configurations in multipotent progenitors. PLoS Genetics. 2014 Jan 9;10(1):e1004079.
15. Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colonystimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proceedings of the National Academy of Sciences. 1997 Jan 21;94(2):569-74.
16. Johansen LM, Iwama A, Lodie TA, Sasaki K, Felsher DW, Golub TR, Tenen DG. c-Myc is a critical target for c/ EBPα in granulopoiesis. Molecular and Cellular Biology. 2001 Jun 1;21(11):3789-806.
17. Hoffman-Liebermann B, Liebermann DA. Suppression of c-myc and c-myb is tightly linked to terminal differentiation induced by IL6 or LIF and not growth inhibition in myeloid leukemia cells. Oncogene. 1991 Jun 1;6(6):903-9.
18. Zakiryanova GK, Shurin MR. Cancer-associated Molecular Abnormalities in Human NK cells. Journal of Cellular Signaling 2021; 2, 94-99.
19. Turkistany SA, DeKoter RP. The transcription factor PU. 1 is a critical regulator of cellular communication in the immune system. Archivum immunologiae et therapiae experimentalis. 2011 Dec 1;59(6):431-40.
20. Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU. 1 in the development of multiple hematopoietic lineages. Science. 1994 Sep 9;265(5178):1573-7.
21. Back J, Allman D, Chan S, Kastner P. Visualizing PU. 1 activity during hematopoiesis. Experimental hematology. 2005 Apr 1;33(4):395-402.
22. Rothenberg EV, Hosokawa H, Ungerbäck J. Mechanisms of action of hematopoietic transcription factor PU. 1 in initiation of T-cell development. Frontiers in Immunology. 2019 Feb 20;10:228.
23. Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU. 1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. Journal of Experimental Medicine. 2005 May 2;201(9):1487-502.
24. DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU. 1. Science. 2000 May 26;288(5470):1439-41.
25. Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU. 1 transcription factors. Immunity. 2006 Nov 1;25(5):731-44.
26. Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007 Oct;26(47):6816-28.
27. van Oevelen C, Collombet S, Vicent G, Hoogenkamp M, Lepoivre C, Badeaux A, et al. C/EBPα activates preexisting and de novo macrophage enhancers during induced pre-B cell transdifferentiation and myelopoiesis. Stem Cell Reports. 2015 Aug 11;5(2):232-47.
28. Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Molecular and Cellular Biology. 1996 Nov;16(11):6083-95.
29. Zhou Y, Wu H, Zhao M, Chang C, Lu Q. The Bach family of transcription factors: a comprehensive review. Clinical Reviews in Allergy & Immunology. 2016 Jun;50(3):345- 56.
30. Kato H, Itoh-Nakadai A, Ebina-Shibuya R, Kobayashi M, Matsumoto M, Muto A, et al. Transcription Factor Bach1 and Bach2 Control Common Myeloid Progenitor Cell Differentiation Under Infectious Stimuli. Blood 2015; 126, 1164-1164.
31. Kato H, Itoh-Nakadai A, Matsumoto M, Ebina- Shibuya R, Sato Y, Kobayashi M, et al. Transcription Factor Bach1 and Bach2 Operate Erythro-Myeloid Competitive Differentiation By Responding to Environmental Changes. Blood. 2016 Jan 1;128(22):2649.
32. Itoh-Nakadai A, Hikota R, Muto A, Kometani K, Watanabe-Matsui M, Sato Y, et al. The transcription repressors Bach2 and Bach1 promote B cell development by repressing the myeloid program. Nature Immunology. 2014 Dec;15(12):1171-80.
33. Roychoudhuri R, Hirahara K, Mousavi K, Clever D, Klebanoff CA, Bonelli M, et al. BACH2 represses effector programs to stabilize T reg-mediated immune homeostasis. Nature. 2013 Jun;498(7455):506-10.
34. Tsukumo SI, Unno M, Muto A, Takeuchi A, Kometani K, Kurosaki T, et al. Bach2 maintains T cells in a naive state by suppressing effector memory-related genes. Proceedings of the National Academy of Sciences. 2013 Jun 25;110(26):10735-40.
35. Nakamura A, Ebina-Shibuya R, Itoh-Nakadai A, Muto A, Shima H, Saigusa D, et al. Transcription repressor Bach2 is required for pulmonary surfactant homeostasis and alveolar macrophage function. Journal of Experimental Medicine. 2013 Oct 21;210(11):2191-204.
36. Igarashi K, Kurosaki T, Roychoudhuri R. BACH transcription factors in innate and adaptive immunity. Nature Reviews Immunology. 2017 Jul;17(7):437-50.
37. Itoh-Nakadai A, Matsumoto M, Kato H, Sasaki J, Uehara Y, Sato Y, et al. A Bach2-Cebp gene regulatory network for the commitment of multipotent hematopoietic progenitors. Cell Reports. 2017 Mar 7;18(10):2401-14.
38. Carvalho JM, Souza MK, Buccheri V, Rubens CV, Kerbauy J, Oliveira JS. CD34-positive cells and their subpopulations characterized by flow cytometry analyses on the bone marrow of healthy allogenic donors. Sao Paulo Medical Journal. 2009;127:12-8.
39. Thom SR, Hampton M, Troiano MA, Mirza Z, Malay DS, Shannon S, et al. Measurements of CD34+/CD45-dim stem cells predict healing of diabetic neuropathic wounds. Diabetes. 2016 Feb 1;65(2):486-97.
40. Ptashne M. On the use of the word ‘epigenetic’. Current Biology. 2007 Apr 3;17(7):R233-6.
41. Jewett A, Kos J, Kaur K, Safaei T, Sutanto C, Chen W, et al. Natural killer cells: diverse functions in tumor immunity and defects in pre-neoplastic and neoplastic stages of tumorigenesis. Molecular Therapy-Oncolytics. 2020 Mar 27;16:41-52.
42. Mognol GP, Spreafico R, Wong V, Scott-Browne JP, Togher S, Hoffmann A, et al. Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proceedings of the National Academy of Sciences. 2017 Mar 28;114(13):E2776-85.
43. Olingy CE, Dinh HQ, Hedrick CC. Monocyte heterogeneity and functions in cancer. Journal of Leukocyte Biology. 2019 Aug;106(2):309-22.
44. Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nature Reviews Cancer. 2016 Jul;16(7):431-46.
45. Zakiryanova GK, Kustova E, Urazalieva NT, Amirbekov A, Baimuchametov ET, Nakisbekov NN, et al. Alterations of oncogenes expression in NK cells in patients with cancer. Immunity, Inflammation and Disease. 2017 Dec;5(4):493-502.
46. Colucci F, Di Santo JP. The receptor tyrosine kinase c-kit provides a critical signal for survival, expansion, and maturation of mouse natural killer cells. Blood, The Journal of the American Society of Hematology. 2000 Feb 1;95(3):984-91.
47. Wang D, Malarkannan S. Transcriptional regulation of natural killer cell development and functions. Cancers. 2020 Jun;12(6):1591.
48. Colucci F, Samson SI, DeKoter RP, Lantz O, Singh H, Di Santo JP. Differential requirement for the transcription factor PU. 1 in the generation of natural killer cells versus B and T cells. Blood, The Journal of the American Society of Hematology. 2001 May 1;97(9):2625-32.
49. Kato H, Itoh-Nakadai A, Matsumoto M, Ishii Y, Watanabe-Matsui M, Ikeda M, et al. Infection perturbs Bach2-and Bach1-dependent erythroid lineage ‘choice’to cause anemia. Nature Immunology. 2018 Oct;19(10):1059- 70.
50. Zhang Z, Liu S, Zhang B, Qiao L, Zhang Y. T cell dysfunction and exhaustion in cancer. Frontiers in Cell and Developmental Biology. 2020 Feb 11;8:17.
51. Zakiryanova GK, Wheeler S, Shurin MR. Oncogenes in immune cells as potential therapeutic targets. ImmunoTargets and therapy. 2018 Apr 11;7:21.
52. Guthrie GJ, Charles KA, Roxburgh CS, Horgan PG, McMillan DC, Clarke SJ. The systemic inflammation-based neutrophil–lymphocyte ratio: experience in patients with cancer. Critical Reviews in Oncology/Hematology. 2013 Oct 1;88(1):218-30.
53. Templeton AJ, McNamara MG, Šeruga B, Vera-Badillo FE, Aneja P, Ocaña A, et al. Prognostic role of neutrophilto- lymphocyte ratio in solid tumors: a systematic review and meta-analysis. JNCI: Journal of the National Cancer Institute. 2014 Jun 1;106(6).
54. Nenclares P, Gunn L, Soliman H, Bover M, Trinh A, Leslie I, et al. On-treatment immune prognostic score for patients with relapsed and/or metastatic head and neck squamous cell carcinoma treated with immunotherapy. Journal for ImmunoTherapy of Cancer. 2021 Jun 1;9(6):e002718.
55. Kohrogi K, Hino S, Sakamoto A, Anan K, Takase R, Araki H, et al. LSD1 defines erythroleukemia metabolism by controlling the lineage-specific transcription factors GATA1 and C/EBPα. Blood advances. 2021 May 11;5(9):2305-18.
56. Kato H, Igarashi K. To be red or white: lineage commitment and maintenance of the hematopoietic system by the “inner myeloid”. Haematologica. 2019 Oct;104(10):1919.
57. Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno SI, Arinobu Y, Geary K, Zhang P, Dayaram T, Fenyus ML. Distinctive and indispensable roles of PU. 1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005 Sep 1;106(5):1590- 600.
58. Etzrodt M, Ahmed N, Hoppe PS, Loeffler D, Skylaki S, Hilsenbeck O, et al. Inflammatory signals directly instruct PU. 1 in HSCs via TNF. Blood, The Journal of the American Society of Hematology. 2019 Feb 21;133(8):816-9.
59. Mossadegh-Keller N, Sarrazin S, Kandalla PK, Espinosa L, Stanley ER, Nutt SL, et al. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 2013 May;497(7448):239-43.
60. Heo SK, Noh EK, Ju LJ, Sung JY, Jeong YK, Cheon J, et al. CD45 dim CD34+ CD38− CD133+ cells have the potential as leukemic stem cells in acute myeloid leukemia. BMC Cancer. 2020 Dec;20(1):1.
61. Heyboer III M, Milovanova TN, Wojcik S, Grant W, Chin M, Hardy KR, Lambert DS, Logue C, Thom SR. CD34+/CD45-dim stem cell mobilization by hyperbaric oxygen—changes with oxygen dosage. Stem Cell Research. 2014 May 1;12(3):638-45.