Commentary
Bio-inspired strategy is kind of interesting to fabricate devices and perform dynamic operations [1]. Various devices have been made, such as airplane, radar and submarine. In life science, as the fundamental entity, million years’ evolution enables cell becomes the most successful functionality. Thus, cell studies undoubtedly deepen our understanding of living systems, and it further bring some predesigned functionalities in living systems [2]. Since Thomas Chang demonstrated the feasibility of artificial cells, scientists have constructed a number of artificial cell-like systems for mimicking cellular molecular signaling [3]. It requires at least three key components towards engineering artificial cell: (i) a metabolic machinery that captures energy and resources for cell living; (ii) a membrane component that keeps the living entity stable and separated from the environment; (iii) a genetic system that processes and transfers information heritance [4,5].
DNA, a genetic molecule, shows great potential in nanotechnology and molecular engineering. Based on DNA hybridization and chain displacement reaction, dynamic DNA nanotechnology shows precisely molecular operations with nanomachines [6], such as DNA logic gate, DNA motor, DNA walker, DNA computing and DNA reaction networks [7]. For structural DNA nanotechnology, topological nanostructures have been designed and constructed by Watson-Crick base pairing, such as DNA dendrimer, DNA framework, DNA hydrogel, and DNA origami [8,9]. In a recent decade, the advancements in DNA nanotechnology inspired us to develop an artificial cell with a DNA engineering way [10]. Several years ago, a biomimetic giant vesicle was generated from detached cancer cells including HeLa and HepG2 cells, which preserved the host cellular properties [11]. Generally, researchers employed synthetic phospholipid bilayer as the membrane models to study cell membrane biology [12]. Compared with the synthetic phospholipid vesicle, our giant vesicle possesses the host cell membrane properties and maintain cellular size. Based on such cell-mimicking giant vesicle, we developed the approach of engineering artificial cell through DNA nanotechnology. We achieved reversible regulation of nanoprisms via DNA hybridization and DNA strand displacement reaction on cell-mimicking surface [13]. Then, a DNA cascade reaction was constructed on cellmimicking surface for mimicking cellular adaptivity [14]. However, the homeostasis of life organism requires a signaling system to transmit information and feedback environmental stimuli. Surface molecular engineering needs to rational interact with the encapsulated system.
In the nature, biomolecular signaling network consist of a series of spatiotemporal and ordered chemical reactions, regulating different biological molecules to maintain living organisms. A key challenge of engineering molecular signaling system is to design and construct a rational integrated functional module to regulate the cascade of molecular events. Therefore, it is attractive to construct a prototype cell with artificial reaction network as the computational core and be able to perform programmed functions [15]. As a proof-of-concept, in February, we reported the new strategies of engineering artificial cell by DNA nanotechnology based on our cellmimicking giant vesicle. A DNA-based artificial molecular signaling system was constructed on our biomimetic giant vesicle, which was derived from a mammalian cell [16]. As shown in Figure 1, a switchable nanochannel was engineered on membrane for materials and information transferring. Given that adenosine triphosphate (ATP) is a canonical energy molecule that triggers cellular machines and drives metabolic reactionsin physiological and pathological reactions, we designed an ATP-responsive DNA nanogatekeeper spanning the membrane, and it serves as the bridge transmitting information between outside and inside of the vesicle. Then, we constructed an encapsulated information processing system. The DNA cascade network was rationally constructed for mimicking cellular signaling pathways. The whole system consists of on-membrane DNA nanostructure and encapsulated DNA signaling network, mimicking the cellular signal reception, transmission and response.
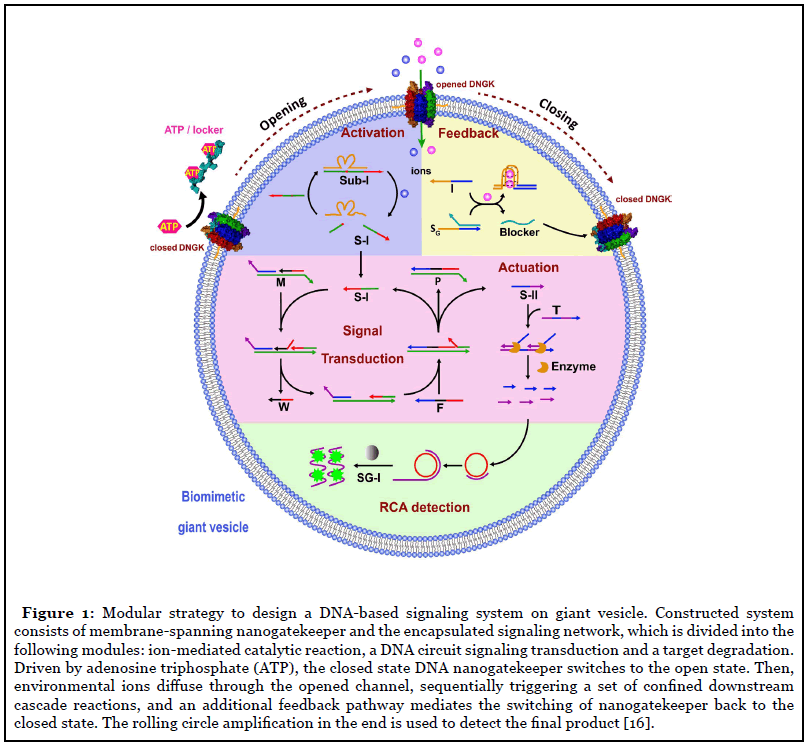
Cellular signaling networks play an important role in cell growth, communication, proliferation, and death. Rapid development of DNA nanotechnology has attracted attention of engineering molecular signaling. In biology, proteins and their interactions play a predominate role in signaling pathway. The strategy of DNA nanotechnology could provide another way to construct an artificial cell. In the future, through precise regulation of DNA nanotechnology, artificial signaling might build powerful applications.
Conflicts of Interest
Authors declare no conflict of interest.
Funding Statement
The authors acknowledge the support from National Postdoctoral Program for Innovative Talents of China (BX20190111).
References
2. Zhu C, Bao G, Wang N. Cell mechanics: mechanical response, cell adhesion, and molecular deformation. Annual Review of Biomedical Engineering. 2000 Aug;2(1):189-226.
3. Chang TMS. Artificial cells. Springfield, IL: Thomas, 1972.
4. Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001 Jan;409(6818):387-90.
5. Ruiz-Mirazo K, Briones C, de la Escosura A. Prebiotic systems chemistry: new perspectives for the origins of life. Chemical Reviews. 2014 Jan 8;114(1):285-366.
6. Bath J, Turberfield AJ. DNA nanomachines. Nature Nanotechnology. 2007 May;2(5):275.
7. Fu T, Lyu Y, Liu H, Peng R, Zhang X, Ye M, et al. DNAbased dynamic reaction networks. Trends in Biochemical Sciences. 2018 Jul 1;43(7):547-60.
8. Lv Y, Peng R, Zhou Y, Zhang X, Tan W. Catalytic selfassembly of a DNA dendritic complex for efficient gene silencing. Chemical Communications. 2016;52(7):1413-5.
9. Seeman NC, Sleiman HF. DNA nanotechnology. Nature Reviews Materials. 2017 Nov 8;3(1):1-23.
10. Noireaux V, Maeda YT, Libchaber A. Development of an artificial cell, from self-organization to computation and self-reproduction. Proceedings of the National Academy of Sciences. 2011 Mar 1;108(9):3473-80.
11. Liu Q, Bi C, Li J, Liu X, Peng R, Jin C, et al. Generating Giant Membrane Vesicles from Live Cells with Preserved Cellular Properties. Research. 2019 Jun 17;2019:6523970.
12. Tian L, Li M, Patil AJ, Drinkwater BW, Mann S. Artificial morphogen-mediated differentiation in synthetic protocells. Nature Communications. 2019 Jul 25;10(1):1-3.
13. Peng R, Wang H, Lyu Y, Xu L, Liu H, Kuai H, et al.Facile assembly/disassembly of DNA nanostructures anchored on cell-mimicking giant vesicles. Journal of the American Chemical Society. 2017 Sep 13;139(36):12410-3.
14. Liu H, Yang Q, Peng R, Kuai H, Lyu Y, Pan X, et al. Artificial signal feedback network mimicking cellular adaptivity. Journal of the American Chemical Society. 2019 Apr 3;141(16):6458-61.
15. Adamala KP, Martin-Alarcon DA, Guthrie-Honea KR, Boyden ES. Engineering genetic circuit interactions within and between synthetic minimal cells. Nature Chemistry. 2017 May;9(5):431.
16. Peng R, Xu L, Wang H, Lyu Y, Wang D, Bi C, et al. DNA-based artificial molecular signaling system that mimics basic elements of reception and response. Nature Communications. 2020 Feb 20;11(1):978.
