Abstract
Introduction: Endometrioid and clear cell carcinomas of the ovary are the most common subtypes of epithelial malignancy arising from endometriosis and are often termed endometriosis-associated ovarian carcinomas (EAOCs). There is a paucity of experimental evidence in the medical literature regarding the role of long non-coding ribonucleic acid (RNA) gene expression in the pathogenesis of these carcinomas.
Purpose: There is a need to develop understanding of the pathogenesis of these carcinomas for neoplastic risk stratification in endometriosis and to develop novel diagnostic biomarkers. Clear cell carcinoma of the ovary, in particular, has a poor prognosis as a result of resistance to standard platinum-based chemotherapy.
Methods: RNAseq datasets from EAOCs were downloaded from Gene Expression Omnibus (GEO) and compared with normal ovarian control sequences using a customized bioinformatic pipeline.
Results: We found 88 differentially expressed non-coding RNA molecules present in both endometrioid and clear cell carcinoma types compared with controls. A further 117 were specifically differentially expressed in the endometrioid carcinoma group and 128 in clear cell carcinoma samples alone. Genes of interest for further study from the 88 shared set in both EAOC types include CASC9, RP4-561L24.3, SLC2A1-AS1, LUCAT1, XIST, CASC15, and MIR99AHG. These genes appear to influence ferroptosis as a common pathway.
Conclusions: Alterations in the ferroptosis pathway may be a key event in development of EAOC in ovarian endometriosis patients. Further work is required to elucidate the function of the candidate RNA genes identified in this study by in-vitro, cell line and cultured organoid experiments. These candidate RNA gene biomarkers have potential clinical utility in early diagnosis, risk stratification of endometriosis, and post-surgical monitoring.
Keywords
Long non-coding RNA, Endometriosis- associated ovarian carcinoma, Pathogenesis, RNA sequencing, Endometrioid adenocarcinoma, Clear cell carcinoma, Ovarian endometriosis
Introduction
Clinical pathology
Ovarian cancer affects 15 women per 100,000 in Europe [1] but it is not one disease. Epithelial malignancy the most common type of ovarian malignancy and defines the groups termed carcinoma [2,3]. Other malignant subtypes include sarcomas, germ cell tumors and sex-cord stromal tumors [1]. Ovarian carcinomas are subdivided based on histological features, the most common being high-grade serous carcinoma which makes up around 70% of ovarian carcinomas [2,4]. Up to 10% of ovarian carcinomas are endometrioid sub-type, having phenotypic and molecular resemblances to endometroid adenocarcinomas that arise in the endometrial cavity [3,5] Clear cell carcinoma (OCCC) and endometrioid carcinoma of the ovary (EnOC) occur on a background of ovarian endometriosis in as many as 70% of cases [6,7]. OCCC is equally as common as endometroid type, perhaps reflecting this shared origin and collectively are often referred to as Endometriosis-Associated Ovarian Carcinomas (EAOCs). Mucinous carcinoma is less common than endometriosis-related carcinomas at around 3% of ovarian carcinomas [2]. Mucinous and low-grade serous carcinomas are rare. Low-grade serous carcinoma has a distinct molecular origin from high-grade serous carcinoma and is regarded as entirely separate entity despite the similarity in their names [2,4,8].
Ovarian endometriosis is a common estrogen dependent disease affecting up to 10% of reproductive-age women around the world [18,19]. It is characterized by the presence of endometrial glands and stroma in sites outside the endometrial cavity [19]. There is an increased lifetime risk of ovarian endometriosis progressing to malignancy of around 1% [20-22]. There is genomic and histological data to suggest that malignancy occurs through an intermediate, dysplastic stage called atypical endometriosis [20,23-26].There is genomic, gene expression, and immunohistochemical evidence to support the theory that endometriosis is a pre-malignant condition [27-32] (Table 1).
|
Histological Diagnosis |
High-grade serous [1,9-12] |
Endometrioid [2,6,12,13] |
Clear cell [2,6,12,13] |
Mucinous [2,12,13] |
Low-grade serous [1,14-17] |
|
Incidence (% of OC) |
70 |
10 |
10 |
3 |
5-10 |
|
Average age at diagnosis |
63 |
56 |
51 |
54 |
47 |
|
Gene mutations |
TP53 BRCA1/2 RAD51C/D BRIP1 MSI genes |
ARID1A, PTEN, CTNNB1, PIK3CA |
ARID1A PIK3CA |
KRAS, HER2 amplification TP53, c-myc |
KRAS NRAS BRAF |
|
Positive IHC |
CK7, ER, WT1, p16, p53, PAX8 |
Vimentin, ER, PR, PAX8 |
CK7, napsinA |
CK7, CK20, cdx2 |
CK7, ER, WT1, PAX8 |
|
FIGO Stage at diagnosis |
51% stage III 29% stage IV |
58-64% stage I |
58-64% stage I |
58-64% stage I |
78% stage I |
|
Platinum-based chemotherapy response |
More than 70% |
60% |
22-56% |
20-60% |
4-40% |
|
Five-year survival |
10-26.9% |
82% |
66% |
71% |
88% |
|
Abbreviations: FIGO: International Federation of Gynecology and Obstetrics; OC: Ovarian Carcinoma; IHC: Immunohistochemistry |
|||||
Gene expression data
It has been shown that increased expression of genes CCNB2, CORO2A, CSNK1G1, FRMD8, LIN54, PDK1, PEX6 and LIN00664 is associated with shorter progression free survival times as compared with serous carcinomas where the converse was observed [33]. This observation reinforces the importance of appropriately segregating ovarian carcinoma subtypes when looking for clinically significant gene expression profiles [33]. Tassi and colleagues found that FOXM1 was differentially expressed between a combined ovarian endometrioid, and clear cell carcinoma group as compared with high-grade serous carcinoma and that this was associated with a poorer prognosis in non-serous carcinoma subtypes [34]. Another tissue microarray study showed over-expression of GLRX, SLC16A3, MKL1, GNE, KIFC3, NAP1, ABCC3, NDRG1, TST, EML2, NP, RAP1GA1, AKR1C1, IGFBP3, ARHB, IMPA2, COL4A2, ANXA4, SLC4A3, FGFR4, TFAP2A, PTPRM, SMTN, ARHGAP8, and C1QTNF6 in OCCC [35]. In contrast, ESR1, ITPR2, WFDC2, FGFRL1, NFIA, SELENBP1, CDH2, PKIB, SCNN1A, IGFBP2, ID4, CMAS, FLOT1, CYP4B1, UBE2F3, GAS1, WT1, EFNB2, MAP1B, DDR1, APOA1B1, TSC22, TRIP7, and EDN1 were under-expressed in the same study [35]. It should be noted that these findings are based on expression data from just six patients. Having said this, high gene expression levels for ANXA4 (annexinA4) and GLRX (glutaredoxin thiotransferase) have been replicated in another study using different experimental techniques [36].
Non-coding RNA
There is emerging data to suggest that elements of the human non-coding genome make a contribution to the pathogenesis of endometriosis-associated ovarian cancers (EAOC) [37]. The non-coding genome plays a part in the development of malignancies across a range of tumor types through transcriptional regulation and control of protein translation by non-coding RNA molecules [38-40].
The RNA molecules responsible for regulating the protein coding genome are divided into long (more than 200 nucleotides) and short molecules (<200 nucleotides). Small RNA molecules include microRNAs (miRNAs) which can direct messenger RNA for degradation before translation. piRNA molecules are PIWI-protein interacting and responsible for silencing transposons in the human genome [41]. Short RNA molecules may be derived from transfer RNA molecules (tsRNA) and these can stabilize messenger RNA for translation in opposition to microRNAs [42]. LncRNA genes play a role in human carcinogenesis by binding to and regulating transcription factors for protein coding genes, inactivating microRNAs that target messenger RNA transcripts for degradation, modifying protein function and cellular localization, influencing chromatin and histone modification, and regulating alternative splicing of mRNA [43]. These functions can affect a number of cell-signaling pathways in the development of cancer including control of cell proliferation, apoptosis and propagating epithelial-mesenchymal transition which is said to confer the ability of epithelial cells to invade connective tissue and metastasize [44-48] (Figure 1).
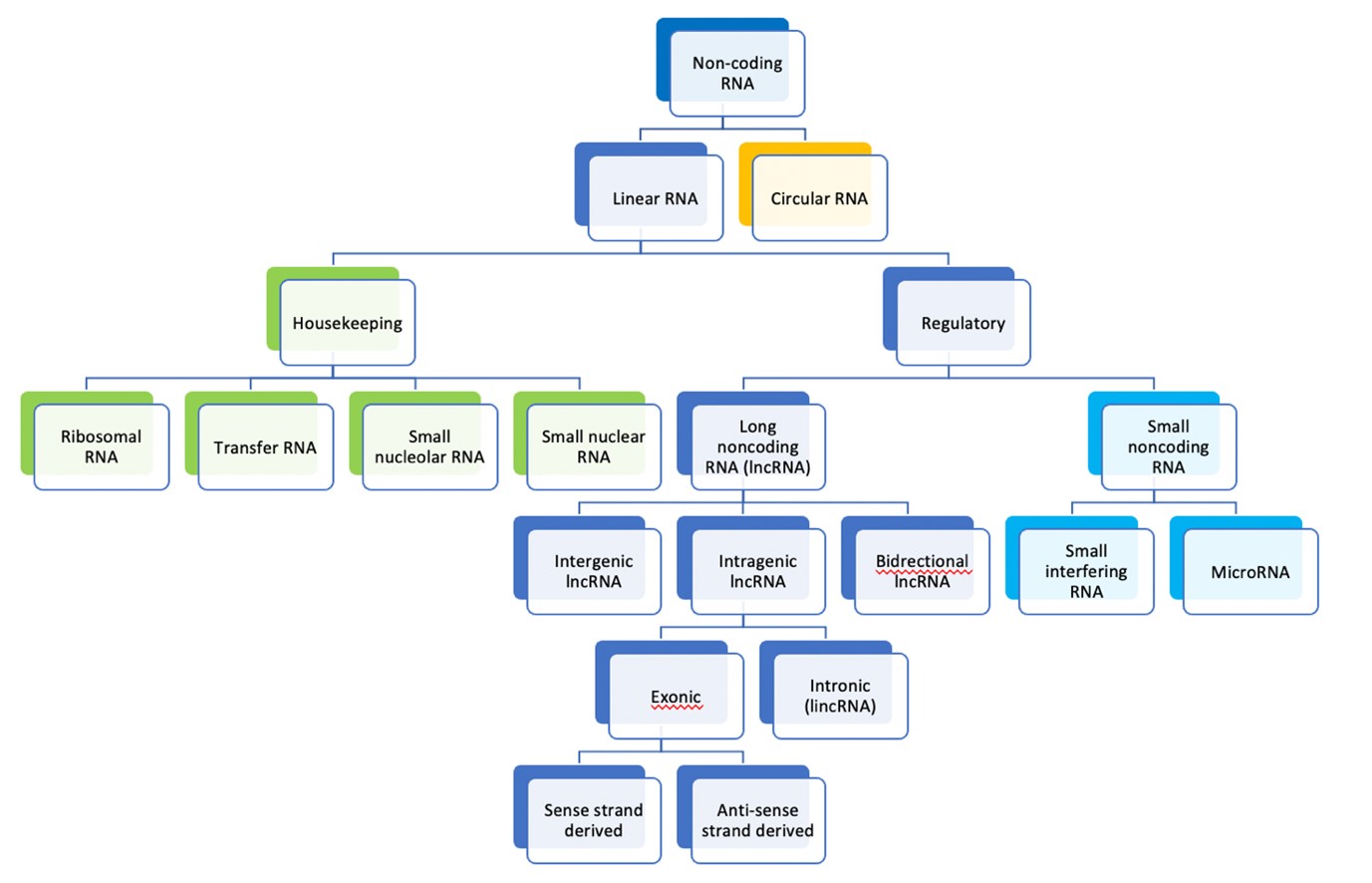
Figure 1. Classification of RNA molecules according to size and cellular function [49].
Long non-coding RNA
It has been shown that many of the non-coding somatic mutations present in EAOC converge on the PAX8 pathway in a range of ovarian cancer subtypes including endometrioid and clear cell subtypes [37]. Endometrial endometrioid adenocarcinoma of the uterine corpus has overlapping molecular pathogenetic characteristics compared with endometrioid adenocarcinoma of the ovary [50]. LncRNA molecule MALAT1 has been shown to be involved in the pathogenesis of endometrioid adenocarcinoma arising from the endometrial cavity by promoting epithelial-mesenchymal transition [51]. NEAT 1, OVAAL, H19, and HOTAIR have also been shown to have altered expression profiles in endometrioid adenocarcinoma [52-56].
Other lncRNA molecules that have been implicated in ovarian carcinogenesis are derived from studies that do not specify the histological subtype of ovarian malignancy. This is largely due to the use of ovarian carcinoma cell lines, most of which derive from high-grade serous carcinoma. The lncRNA genes differentially expressed in cell lines include ANRIL, BC200, HULC, HST2, HOST2, GAS5, PTAF, SOX2OT, DGCR5, PC3A, FAL1, ABO73614, LSINCT5, PVT1, LINK-A, HOXA11-AS, PVT1, TUG1, UCA1, ZFAS1, the majority of which are said to behave as oncogenes [56-61].
This study is a meta-analysis of published RNA sequencing (RNA-seq) data sets generated through high-throughput sequencing methods for differential expression analysis using a customized bioinformatics pipeline. The aim was to document the differential gene expression profile of EAOC with focus on lncRNA genes. Secondarily, the function and pathway involvement of these lncRNA genes was to be sought from in silico tools and databases for insights into EAOC pathogenesis.
Methods
This study is a meta-analysis of data generated by RNA sequencing by other researchers posted in a public access online database for the purpose of further analysis.
Data set identification
An online search of the gene expression omnibus (GEO) database as curated by NCBI (National Center for Biotechnology Information) was performed using keywords ovary, endometriosis-associated ovarian carcinoma, ovarian cancer, atypical endometriosis, endometriosis, clear cell carcinoma and ovarian endometrioid carcinoma. The resulting search results were filtered using the terms ‘Homo sapiens’, and ‘expression profiling by high throughput sequencing’. Tissue samples from ovarian carcinomas other than EAOCs, metastatic disease, fetal and embryonic tissues, fluid cytology samples, stem cells, circulating tumour cells and cell lines were excluded. Application of exclusion criteria yielded 1960 human RNA-seq data sets; five were for normal ovarian tissue (Geo Accession numbers GSM1010948, GSE127873, GSE137608, GSE135485, GSE18927), four were for endometriosis-related controls (eutopic and ectopic endometrial tissues and normal endometrium from healthy patients; accession numbers GSE118928, GSE99949, GSE87809, GSE87810), and one for EAOC samples (GSE121103). There were no RNA-seq data sets available that included atypical endometriosis samples.
Clinical details of ovarian carcinoma tissue samples
Patient information for each of the source samples used to generate the RNA-seq data for GSE121103 is given in Table 2. One of the endometrioid carcinoma samples (EnOC) failed to generate reads of sufficient quality for publication to GEO but the investigators do not specify which of the samples this refers to in their paper [37].
|
|
Age |
Ethnicity |
Tumour grade |
Tumour stage |
|
OCCC 1 |
45 |
Asian |
3 |
IIIC |
|
OCCC 2 |
47 |
White Hispanic |
3 |
IIIC |
|
OCCC 3 |
61 |
Unknown |
3 |
IIIC |
|
OCCC 4 |
38 |
Unknown |
3 |
IIIB |
|
OCCC 5 |
52 |
Unknown |
3 |
IIIB |
|
EnOC 1 |
64 |
Hispanic |
3 |
IV |
|
EnOC 2 |
50 |
White Hispanic |
1 |
IB |
|
EnOC 3 |
35 |
White Hispanic |
2 |
IC |
|
EnOC 4 |
42 |
White Hispanic |
2 |
IC |
|
EnOC 5 |
41 |
White Hispanic |
1 |
IC |
|
Abbreviations: OCCC: Ovarian Clear Cell Carcinoma; EnOC: Endometrioid adenocarcinoma of the Ovary. |
||||
Bioinformatic pipeline
Fastq files are first assessed for quality using fastQC [62]. The RNA sequencing reads were aligned to the reference genome using STAR aligner which generates a gene count table file labelled ReadsPerGene.out.tab. The resulting alignment from STAR generates a file of summary mapping statistics annotated as Log.final.out. This gives an indication of the quality of the sample analyzed by reflecting the proportion of input reads, the average RNA molecule read length versus the number of unmapped and chimeric reads and mismatch rate.
A data matrix of factors informed the normalization step which was carried out by using the DESeq2 package. DESeq2 adjusts for the variation in expression counts according to variation in the read depth for each gene by using a generalized linear model and creates an estimate of moderated variance of genes by comparing the variance of the gene in question versus the average variance of all the genes present in the dataset [63]. Following normalization, DESeq2 performs differential expression analysis and calculates the probability of the difference seen having occurred by chance as expressed as a p value. Statistical significance was set at the 0.05 level. The gene lists were filtered for base mean expression (absolute expression level) >10 and log fold change >2 (i.e. at least doubling of under or over-expression).
Biological interpretation
Online bioinformatics tools and databases, including NCBI, Ensembl, OMIM, Clinvar, Genecards, RISE, Diana, and PubMed were used to identify function and intra-cellular pathway influence. Ensembl was used to identify nearest protein coding genes to the lncRNA genes identified. RNA databases RNAcentral, Rfam, NONCODE, LNCipedia, LNCBook, and lncrnadb were used to provide up-to-date information regarding all aspects of lncRNA genes identified.
Ethical Considerations
Meta-analysis was chosen as the method of study, in part, due to the temporal constraints on ethical review but also because of financial limitations. Specific ethical approval was not required for this study as it was a re-analysis of published patient sequencing data in the public domain. The tissue samples used in the original study by Corona et al., 2018 were collected with informed consent and given approval by the ethical review board of the University of Southern California.
Results
The final read out sheets for all samples in Appendix 2 show give an overall summary of nucleic acid quality prior to analysis in the bioinformatic pipeline. The samples for normal endometrium were of insufficient quality to use as control material for this study. The final gene list for the endometrial samples show read out counts of zero across all gene identifiers for all three RNA extraction samples of endometrium. Normal ovarian tissue was therefore used as control material as sequencing read outs were of good quality.
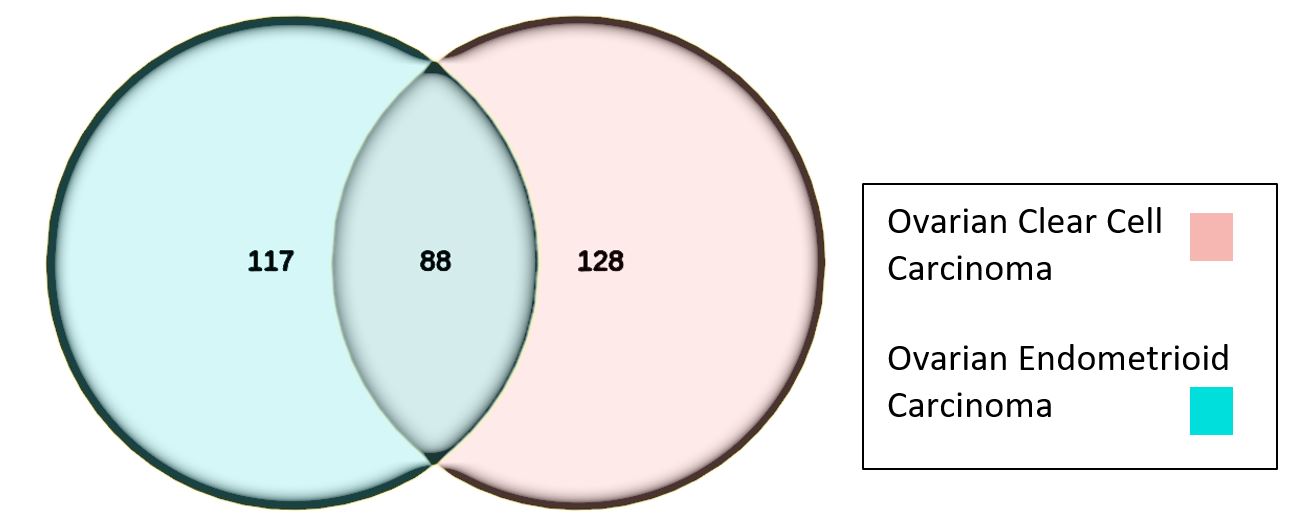
The primary aim of describing the differential gene expression of EAOCs was achieved. The secondary aim of functional characterization was also achieved but required assumption of in cis function of all lncRNA genes to inform interpretation. A total of 35,697 genes were differentially expressed in the ovarian endometrioid adenocarcinoma (EnOC) sample set from 4 patients (n=4) and 33,939 genes from the ovarian clear cell carcinoma (OCCC) samples (n=5). Both gene expression lists were filtered by removing all protein coding genes, microRNAs (less than 200 nucleotides in length), processed transcripts, pseudogenes (processed and unprocessed), small nuclear RNA (snRNA) and small nucleolar (snoRNA), mitochondrial RNA and molecules classified as miscellaneous. The total gene expression list included 333 lncRNA genes with 88 being present in both the endometrioid and clear cell adenocarcinoma groups. There was differential expression of 117 genes in the endometrioid group alone and 128 differentially expressed genes in the clear cell group (Figure 2).
Figure 2. Venn diagram illustrating number of overlapping lncRNA molecules between groups of ovarian endometriosis-related adenocarcinomas (filtered by LFC >2, BM>10 and p<0.05).
The top ten most over expressed genes, in decreasing order, in the overlapping group of 88 lincRNA genes were RP11-456B22.8, LINC00958, LINC00621, RP11-529E15.1, RP11-3J1.1, RP11-4K16.2, LINC01320, U47924.27, CASC9, and RP1-86C11.7 as measured by absolute log fold change (LFC) >2.
The most under-expressed lincRNA genes in the overlapping group compared with control samples were RP4-561L24.3, RP11-108M9.3, AC084082.3, RP4-535B20.1, CTD2332E11.2, RP11-473M20.16, LINC00324, RP11-613D13.8, AP001172.3, and RP5-875O13.1, with RP4-561L24.3 being the most under-expressed with a LFC of -11.29. See Tables 3 and 4.
|
Gene name |
Expression level |
Log Fold Change |
||
|
|
EnOC |
OCCC |
EnOC |
OCCC |
|
RP11-456B22.8 |
38.41 |
45.80 |
9.53 |
10.23 |
|
LINC00958 |
92.48 |
248.13 |
10.72 |
9.62 |
|
LINC00621 |
399.39 |
142.08 |
9.83 |
9.46 |
|
RP11-529E15.1 |
30.39 |
30.08 |
8.91 |
8.95 |
|
RP11-3J1.1 |
39.98 |
30.85 |
8.25 |
8.76 |
|
RP11-4K162 |
26.01 |
19.70 |
8.46 |
8.51 |
|
LINC01320 |
624.39 |
596.55 |
7.66 |
8.39 |
|
U47924.27 |
23.19 |
21.39 |
7.32 |
7.95 |
|
CASC9 |
50.44 |
82.20 |
7.84 |
7.55 |
|
RP1-86C11.7 |
12.28 |
14.06 |
7.83 |
7.39 |
|
|
|
|
|
|
|
RP4-561L24.3 |
3555.92 |
2716.14 |
-11.87 |
-11.29 |
|
RP11-108M9.3 |
556.1 |
424.11 |
-11.22 |
-9.93 |
|
AC084082.3 |
240.31 |
181.83 |
-8.22 |
-7.26 |
|
RP4-535B20.1 |
29.40 |
21.43 |
-8.23 |
-6.04 |
|
CTD-2332E11.2 |
140.74 |
107.36 |
-5.85 |
-5.62 |
|
RP11-473M20.16 |
229.46 |
172.49 |
-6.03 |
-5.38 |
|
LINC00324 |
230.15 |
169.80 |
-6.37 |
-5.33 |
|
RP11- 613D13.8 |
156.39 |
115.61 |
-5.82 |
-5.12 |
|
AP001172.3 |
18.88 |
15.71 |
-5.56 |
-4.87 |
|
RP5-875O13.1 |
28.88 |
21.69 |
-4.77 |
-4.51 |
|
Abbreviations: EnOC: Ovarian Endometrioid adenocarcinoma; OCCC: Ovarian Clear Cell Carcinoma. Colour code: Red = Over-expression; Green = Under-expression. |
||||
The most differentially overexpressed lincRNA molecules in the endometrioid carcinoma group were RP11-6.08O21.1, AC011288.2, LINC01123, LLINC01508, RP11-400N13.2, RP11-319E16.2, LINC010206, RP1-60O19.1, CTC-304I17, and RP11-89K21.1 whilst those with most reduced expression out of the 117 lincRNA molecules identified were much fewer in number. They are RP11-1100L3.8, GATA6-AS1, RNU12, RP11-323I15.5, LINC00602, and RP11-95H3.1.
|
Gene Name |
Expression level |
Log fold change |
P value |
|
RP11-608O21.1 |
35.93 |
9.15 |
1.67E-08 |
|
AC011288.2 |
27.25 |
9.11 |
9.16E-07 |
|
LINC01123 |
29.45 |
8.78 |
2.49E-07 |
|
LINC01508 |
22.18 |
8.68 |
8.71E-07 |
|
RP11-400N13.2 |
17.80 |
8.44 |
6.73E-06 |
|
RP11-319E16.2 |
20.7 |
8.32 |
3.69E-05 |
|
LINC01206 |
21.03 |
8.0 |
4.06E-05 |
|
RP1-60O19.1 |
16.41 |
8.02 |
4.48E-05 |
|
CTC-304I17.6 |
18.01 |
7.83 |
4.66E-05 |
|
RP11-89K21.1 |
83.85 |
7.81 |
3.04E-09 |
|
|
|
|
|
|
RP11-1100L3.8 |
90.92 |
-2.07 |
0.0016 |
|
GATA6-AS1 |
68.26 |
-2.65 |
0.0002 |
|
RNU12 |
13.14 |
-2.6 |
0.005 |
|
RP11-323I15.5 |
15.44 |
-2.70 |
0.004 |
|
LINC00602 |
12.94 |
-3.73 |
0.002 |
|
RP11-95H3.1 |
48.37 |
-3.74 |
8.05E-05 |
|
Red = Over-expression; Green = Under-expression |
|||
Of the 128 lincRNA molecules found in the clear cell carcinoma group the following were most over-expressed: LINC00668, LINC00858, RP11-190J1.3, LINC01446, LINC01518, RP11-528A4.2, RP11-356C4.5, LINC01559, CTD-2008P7.8, RP11-346C4.3. The greatest reduction in expression compared with control normal ovary included ENOX1-AS1, AP000962.2, OVAAL, RP11-400K9.4, RP11- 1081M.51, LINC01539, LINC00924, RP11-826N14.4, GAS1RR, and LINC01018 (Tables 5 and 6).
|
Gene Name |
Expression level |
LFC |
P value |
|
LINC00668 |
508.97 |
11.72 |
1.23E-20 |
|
LINC00858 |
35.23 |
9.63 |
8.86E-09 |
|
RP11-190J1.3 |
38.88 |
9.63 |
3.54E-08 |
|
LINC01446 |
24.76 |
8.79 |
8.23E-05 |
|
LINC01518 |
30.61 |
8.26 |
1.58E-06 |
|
RP11-528A4.2 |
18.89 |
7.91 |
5.62E-05 |
|
RP11-356C4.5 |
11.15 |
7.61 |
8.16E-05 |
|
LINC01559 |
102.25 |
7.49 |
4.19E-05 |
|
CTD-2008P7.8 |
11.03 |
7.22 |
0.00034 |
|
RP11-346C4.3 |
22.28 |
6.96 |
7.43E-05 |
|
|
|
|
|
|
ENOX1-AS1 |
13.79 |
-5.26 |
2.32E-05 |
|
AP000962.2 |
35.17 |
-5.82 |
1.71E-05 |
|
OVAAL |
10.51 |
-6.16 |
0.0013 |
|
RP11-400K9.4 |
45.77 |
-6.29 |
1.97E-08 |
|
RP11- 1081M.51 |
10.16 |
-6.32 |
0.00056 |
|
LINC01539 |
14.00 |
-6.40 |
0.0020 |
|
LINC00924 |
71.39 |
-6.90 |
3.01E-10 |
|
RP11-826N14.4 |
13.56 |
-6.90 |
0.00032 |
|
GAS1RR |
29.61 |
-6.92 |
2.16E-06 |
|
LINC01018 |
31.93 |
-6.99 |
1.18E-06 |
|
Red = Over-expression; Green = Under-expression |
|||
|
Endometrioid Adenocarcinoma |
Both CCC and Endometrioid |
Clear Cell Carcinoma (CCC)
|
|||
|
LncRNA greatest log fold change |
lncRNA highest expression level |
LncRNA greatest log fold change |
lncRNA highest expression level |
LncRNA greatest log fold change |
lncRNA highest expression level |
|
lncSLIT2 SLIT2 |
LINC01695 N6AMT1 |
RP11-456B2 2.8 RNF223 |
RP4-561L24.3 BCAR3 GCLM DNTTIP2 |
LINC00668 LAMA1 ARHGAP28 |
XIST TSIX HIF1A |
|
AC011288.2 ARL4A |
RP11-191L9.4 TBC1D22A |
LINC00958 TEAD1
|
CARMN PCYOX1L |
LINC00858 LRIT1 RGR |
C1orf132 CD34 CD46 |
|
LINC01123 MALL |
BLACAT1 LEMD1 |
LINC00621 SGCG
|
SLC2A1-AS1 SLC2A1 HIF-1alpha |
RP11-190J1.3 FBXW4 |
LINC00668 LAMA1 ARHGAP28 |
|
LINC01508 DIRAS2 |
LUCAT1 ADGRV1 NRF2 |
RP11-529E15.1 FAM98A |
LINC00478 USP25
|
LINC01446 VS2MTA POM121L12 |
MIRLET7BHG PRR34 |
|
LINC02474 DUSP10 |
LINC02604 TMEM248
|
RP11-3J1.1 LCORL SLIT2 |
PWRN1 NPAP1 |
LINC01518 ZNF338 |
RP11-54H7.4 MYO16 |
|
LNCNFT53-2 NFT3 |
NRAD1 LACC1 CCDC122 |
MAL2-AS1 MAL2 |
CASC15 PRL SOX4 CDKAL1 |
LINC02038 OPA1 HES1 |
RP11-20D14.6 RIMKLB |
|
LINC01206 SOX2 |
KRT80-4 NR4A1 |
LINC01320 FAM98A |
CH507-513H4.6 KCNE1B
|
RP11-356C4.5 PRDM7 |
HCG11 BNT1A1 HMGN4 |
|
RP1-60O19.1 PDSS2 |
RP-11-89K21.1 |
MIR200CHG PHB2 |
LINC01320 FAM98A |
LINC01559 GRIN2B
|
MIR29A KLF4 MKLN1 |
|
CTC-304I17.6
|
LINC00937 |
CASC9 HN4A |
RP11-108M9.3 |
CTD-2008P7.8 |
RP11-554D15.3 |
|
LncRNA genes are in black, adjacent protein coding genes identified in Ensembl are in blue. Genes involve in ferroptosis are shown in red. |
|||||
Discussion
Long non-coding RNAs have several modes of function in human cells and these fall into three broad categories; post-mRNA processing, chromatin reprogramming and regulation of protein coding gene transcription and enhancer sites [64]. Many lncRNA molecules are said regulate transcription factors of nearby protein coding gene and are thus cis-acting [65-68]. There is a paucity of published literature regarding lncRNA function with mRNA and miRNA interactions for the majority of lncRNA genes listed in the results above. Understanding that many long non-coding RNA molecules function in cis allowed detailed exploration of the genomic sites of origin of these molecules and generation of a list of protein coding genes that represent the most likely targets to be controlled by the lncRNAs identified. Analysis of these genes with long-non-coding RNA species in the Genecards database has identified potential molecular function and highlighted biological pathways in which they function. For example, lncRNA cancer susceptibility 9 (CASC9) is situated next to the hepatocyte nuclear factor 4 gamma (HNF4G) protein coding gene (also known as NR2A2).
OVAAL (ovarian adenocarcinoma amplified lncRNA) expression has been reported as amplified in ovarian high-grade serous carcinoma [69]. In this study, OVAAL showed reduced expression in ovarian clear cell carcinoma (log fold change -6.16, p=0.001). Research suggests that OVAAL behaves as an oncogene by enhancing cell survival through initiation of the RAF/MEK/ERK pathway and avoiding cellular senescence mechanisms (70). However, down-regulated expression of OVAAL in this dataset appears to contradict this evidence and would suggest a tumor suppressor function in vivo.
Findings by Zou et al., 2015 support the hypothesis that CASC9 interacts with protein coding HNF4G by acting in cis based on evidence from datamining bioinformatic online databases [71]. HNF4G is one of a subfamily of liver-specific transcription factors important for organ development in utero [72]. HNF4 has also been shown to be overexpressed in Islet of Langerhans cells of the pancreas in young people with maturity onset diabetes [73]. The morphological feature of cytoplasmic clearing in OCCC is due to intracytoplasmic glycogen accumulation and this may be due to alterations in glycogen metabolism as a result of alterations in HNF4G function. HNF4G has been documented as being involved in OCCC pathogenesis [37]. Also, strong immunohistochemical expression of a different subset of HNF, HNF1beta, has been shown in OCCC but this is not used in routine diagnostic histopathological practice due to a lack of specificity and distinct morphology [74]. Furthermore, the genes for the group of transcription factors comprising HNF4 and HNF1 are found on different chromosomes and activate transcription of different cytochrome p450 enzymes [75].
Cdc42 interacts with breast cancer anti-oestrogen resistance protein 3 (BCAR3) which interacts with RP4-561L24.3. Cdc42 codes for a cell membrane protein found in macrophages and is responsible, in part, for coordinated and effective phagocytosis [76]. Reduced expression of cdc42 protein on the surface of macrophages has been shown to differ between tissues with endometriosis and EAOC. Loss of cdc42 expression may play a role in the malignant transformation of endometriosis. Further, cdc42 protein also plays a key role in the MAPK pathway that controls cellular proliferation, anti-apoptosis and cellular differentiation.
In addition, BCAR3, RP4-561L24.3 is co-located with GCLM and DNTTIP2 on chromosome 1 [77]. In this meta-analysis, RP4-561L24.3 was expressed at the highest overall level in the combined EAOC group but simultaneously differentially under-expressed in comparison with controls. Ferroptosis is a form of programmed cell death resulting from a combination of iron and lipid peroxidation in mitochondria with a toxic accumulation of reactive oxygen species [78]. Resistance to ferroptosis is thought to be critical development in the pathogenesis of endometriosis [79]. This makes biological sense in a context of endometriosis where the local tissue environment will contain increased amounts of iron-rich haemosiderin as a consequence of menstrual bleeding. However, the role of ferroptosis in the development of endometriosis-associated malignancy has yet to be described in full. A recent report showed an increased sensitivity of EAOCs to Erastin therapy, which targets the ferroptosis pathway, leads to increased rates of cell death [80]. If resistance to ferroptosis is an important mechanism in the development of EAOC, one could hypothesize that gene expression studies will show increased expression of tumor suppressor genes in normal cells with reduced differential expression in malignant cells [81,82]. This suggests that RP4-561L24.3 functions as a tumor suppressor as it is the most down regulated lncRNA molecule in this EAOC dataset in the combined endometrioid and clear cell group (log fold change -11.87, p value= 3.97x 10-34).
Further evidence of a role for ferroptosis in the development of EAOC comes from the finding that LUCAT1 controls NRF2 expression via miRNA-495 [83]. NRF2 (also known as NFE2L2) has been shown to have a critical role in ferroptosis [84]. NRF2 (Nuclear-Factor Erythroid2-Related Factor) is a transcription factor that regulates genes with promotors containing anti-oxidant response elements [85]. Genes regulated by NRF2 are upregulated in response to oxygen free radicals released in a context of inflammation and injury as seen in ovarian tissues affected by endometriosis [86].
It is also interesting to note that cells with high levels of nrf2 protein activity are enriched for gamma-glutamyl peptides which are produced as a result of GCLC/GCLM activity [87]. Kang, et al. suggest that GCLM may be regulated in cis by RP4-561L24.3. It would make sense that reduced differential expression of genes known to act in the ferroptosis pathway play a key role in EAOC pathogenesis as it is known that menstrual cycling in endometriotic lesions results in generation of excessive irons and free radicals [80].
SLC2A1 is a member of the family of solute carriers. The anti-sense lncRNA molecule SLC2A1-AS1 is expressed at a high level in our EAOC data. This regulates transcription of SLC2A1 on the sense strand of DNA (88). Transcription of SLC2A1 is induced under hypoxic conditions and influenced by increasing levels of hypoxia inducible factor 1 alpha (HIF-1alpha) [89].
SOX4 is said to be involved in the pathway regulating ferroptosis [90,91]. This may be regulated in cis by CASC15 due to co-location with SOX4 in the human genome [77].
SOX4 (SRY-related HMG-box 4) is a transcription factor that has been implicated in carcinogenesis as high expression of SOX4 is thought to contribute to dedifferentiation, cell survival and epithelial-mesenchymal transition [92]. On the basis of its molecular interactions, CASC15 may have an important role to play in development of EAOC. Researchers suggest that CASC15 functions as a tumor suppressor gene [93] in which case one would expect to see reduced levels of expression compared with controls in this study. This is not the case, however, as CASC15 was shown to be expressed at a high level and was not differentially expressed. This casts some doubt on the role of CASC15 in the development of EAOC and requires further experimental investigation.
Mir-99a-Let7c Cluster Host Gene (MIR99AHG), otherwise known as LINC00478, resides near to protein coding gene Ubiquitin Specific Peptidase 25 (USP25). USP25 is a peptidase enzyme responsible for cleavage of ubiquitin from proteins destined for cellular degradation by proteolysis in the proteosome [94]. It thereby prevents target proteins from breakdown and has been shown to interact with components of the MAPK pathway [95]. USP25 also inhibits induction of ferroptosis in malignancy [96].
XIST has been documented as having an influence on ferroptosis by suppressing glutathione-S transferase (GST) and increasing glutathione synthase levels [97]. XIST was expressed at the highest overall level in OCCC but was simultaneously down regulated (expression level 5357, log fold change -2.68, p value= 0.0001. There is evidence in the RISE database of RNA-RNA interactions that XIST interacts with anti-sense RNA HIF1alpha-AS1 which regulates transcription of the hypoxia inducible factor from the opposite coding strand of DNA [98]. HIF1alpha is also induces expression of SCL2A1-AS1 under hypoxic conditions [89] (Figure 3).
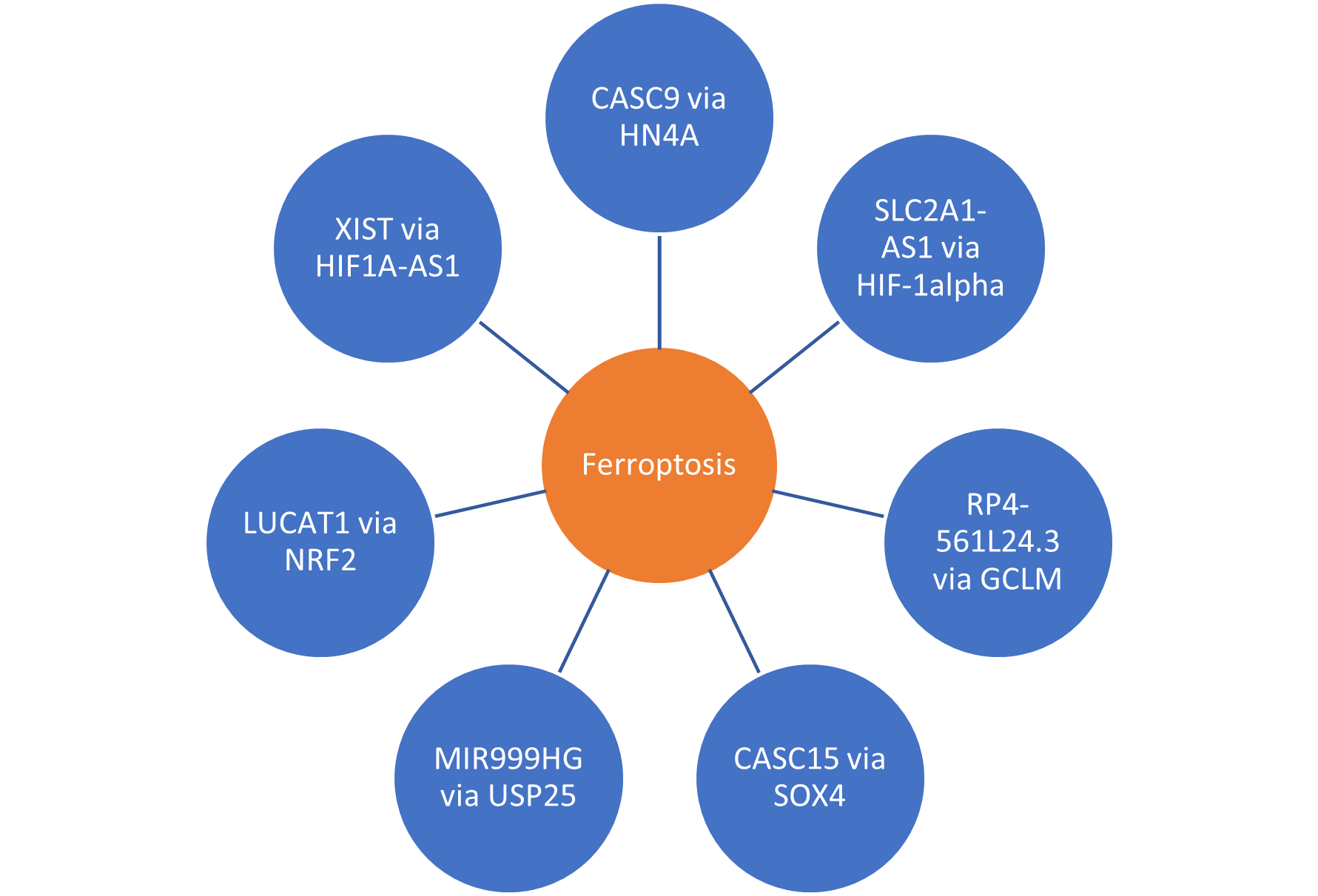
Figure 3. Summary of lncRNA molecules converging on ferroptosis.
Strengths and Limitations of This Study
Limitations of this study include not having access to the source tissue to confirm the disease entities listed in the patient clinical information table were histologically accurate. There is concern regarding the specimen classified as FIGO grade 3 endometrioid adenocarcinoma of the ovary as this is a difficult histological diagnosis to make. The morphology of grade 3 endometrioid adenocarcinoma is often solid, poorly differentiated, and similar to high-grade serous carcinoma. The distinction relies upon the use of a panel of immunohistochemistry markers which were not described in original publication [37].
Another limitation of the study was the use of normal ovarian tissue as a source of control RNA sequence for comparison with the malignant tissues. It would have been more accurate to use ovarian endometriosis samples given that this is said to be the cell of origin for most EAOCs [6] but these were of insufficient quality for use.
The online databases used to study the pathways are at an early stage in evolution with gaps in knowledge content. In many cases there is no known functional information or interactions regarding the lncRNA species identified in this meta-analysis. Furthermore, many of the assertions made are based on in silico prediction rather than experimental data. This indicates a need for further work by, for example, performing knockdown experiments in organoid models of endometriosis and EAOCs to help classify function and contribute to the knowledge base. A major assumption of the interpretation of our data rests on a cis- rather than trans-regulatory function of the lncRNA molecules identified in this work [66].
Conclusions
LncRNA genes RP4-561L24.3, CASC15, CASC9, SLC2A-AS1, LUCAT1, XIST and MIR99AHG are candidate biomarkers for further exploration in the pathogenesis of EAOC. Some of these lncRNA molecules may have an influence on the ferroptosis pathway by cis-acting regulation of transcription factors of nearby protein coding genes [66]. In vitro experiments are required to provide evidence in respect of this hypothesis. Numerous online databases have incomplete information relating to lncRNA function and interactions that makes further experimentation necessary.
Ferroptosis is a form of programmed cell death that is induced in response to cellular stresses involving iron and lipid metabolism in mitochondria and might play a part in the pathogenesis of EAOC. LncRNA molecules identified in this meta-analysis may inform clinical diagnostic pathways by development of non-invasive methods of detection for early diagnosis, risk stratification of endometriosis and more effective post-surgical patient monitoring for recurrence.
Conflicts of Interest
The authors declare no conflicts of interest.
References
2. Prat J, D'Angelo E, Espinosa I. Ovarian carcinomas: at least five different diseases with distinct histological features and molecular genetics. Hum Pathol. 2018;80:11-27.
3. Kaku T, Ogawa S, Kawano Y, Ohishi Y, Kobayashi H, Hirakawa T, et al. Histological classification of ovarian cancer. Med Electron Microsc. 2003;36(1):9-17.
4. McCluggage WG. Morphological subtypes of ovarian carcinoma: a review with emphasis on new developments and pathogenesis. Pathology. 2011;43(5):420-32.
5. Wu RC, Veras E, Lin J, Gerry E, Bahadirli-Talbott A, Baras A, et al. Elucidating the pathogenesis of synchronous and metachronous tumors in a woman with endometrioid carcinomas using a whole-exome sequencing approach. Cold Spring Harb Mol Case Stud. 2017;3(6).
6. McCluggage WG. Endometriosis-related pathology: a discussion of selected uncommon benign, premalignant and malignant lesions. Histopathology. 2020;76(1):76-92.
7. Wang Y, Mang M, Wang L, Klein R, Kong B, Zheng W. Tubal origin of ovarian endometriosis and clear cell and endometrioid carcinoma. Am J Cancer Res. 2015;5(3):869-79.
8. Muinao T, Pal M, Deka Boruah HP. Origins based clinical and molecular complexities of epithelial ovarian cancer. International Journal of Biological Macromolecules. 2018;118(Pt A):1326-45.
9. Taylor A, Brady AF, Frayling IM, Hanson H, Tischkowitz M, Turnbull C, et al. Consensus for genes to be included on cancer panel tests offered by UK genetics services: guidelines of the UK Cancer Genetics Group. J Med Genet. 2018;55(6):372-7.
10. Singh N, McCluggage WG, Gilks CB. High-grade serous carcinoma of tubo-ovarian origin: recent developments. Histopathology. 2017;71(3):339-56.
11. UK CR. Ovarian Cancer Survival Statistics by Stage 2019 [
12. Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284-96.
13. Chávarri-Guerra Y, González-Ochoa E, De-la-Mora-Molina H, Soto-Perez-de-Celis E. Systemic therapy for non-serous ovarian carcinoma. Chin Clin Oncol. 2020.
14. Kaldawy A, Segev Y, Lavie O, Auslender R, Sopik V, Narod SA. Low-grade serous ovarian cancer: A review. Gynecol Oncol. 2016;143(2):433-8.
15. Schmeler KM, Sun CC, Bodurka DC, Deavers MT, Malpica A, Coleman RL, et al. Neoadjuvant chemotherapy for low-grade serous carcinoma of the ovary or peritoneum. Gynecol Oncol. 2008;108(3):510-4.
16. Dalton HJ, Fleming ND, Sun CC, Bhosale P, Schmeler KM, Gershenson DM. Activity of bevacizumab-containing regimens in recurrent low-grade serous ovarian or peritoneal cancer: A single institution experience. Gynecol Oncol. 2017;145(1):37-40.
17. Slomovitz B, Gourley C, Carey MS, Malpica A, Shih IM, Huntsman D, et al. Low-grade serous ovarian cancer: State of the science. Gynecol Oncol. 2020;156(3):715-25.
18. Somigliana E, Vigano' P, Parazzini F, Stoppelli S, Giambattista E, Vercellini P. Association between endometriosis and cancer: a comprehensive review and a critical analysis of clinical and epidemiological evidence. Gynecol Oncol. 2006;101(2):331-41.
19. Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364(9447):1789-99.
20. Sainz de la Cuesta R, Eichhorn JH, Rice LW, Fuller AF, Jr., Nikrui N, Goff BA. Histologic transformation of benign endometriosis to early epithelial ovarian cancer. Gynecol Oncol. 1996;60(2):238-44.
21. Wei JJ, William J, Bulun S. Endometriosis and ovarian cancer: a review of clinical, pathologic, and molecular aspects. Int J Gynecol Pathol. 2011;30(6):553-68.
22. Dawson A, Fernandez ML, Anglesio M, Yong PJ, Carey MS. Endometriosis and endometriosis-associated cancers: new insights into the molecular mechanisms of ovarian cancer development. Ecancermedicalscience. 2018;12:803.
23. Jiang X, Morland SJ, Hitchcock A, Thomas EJ, Campbell IG. Allelotyping of endometriosis with adjacent ovarian carcinoma reveals evidence of a common lineage. Cancer Res. 1998;58(8):1707-12.
24. Eurich KE, Goff BA, Urban RR. Two cases of extragonadal malignant transformation of endometriosis after TAH/BSO for benign indications. Gynecol Oncol Rep. 2019;28:23-5.
25. Kajiyama H, Suzuki S, Yoshihara M, Tamauchi S, Yoshikawa N, Niimi K, et al. Endometriosis and cancer. Free Radic Biol Med. 2019;133:186-92.
26. Dentillo DB, Meola J, Ferriani RA, Rosa-E-Silva JC. Common Dysregulated Genes in Endometriosis and Malignancies. Rev Bras Ginecol Obstet. 2016;38(5):253-62.
27. Herreros-Villanueva M, Chen CC, Tsai EM, Er TK. Endometriosis-associated ovarian cancer: What have we learned so far? Clin Chim Acta. 2019;493:63-72.
28. Lac V, Verhoef L, Aguirre-Hernandez R, Nazeran TM, Tessier-Cloutier B, Praetorius T, et al. Iatrogenic endometriosis harbors somatic cancer-driver mutations. Human Reproduction. 2019;34(1):69-78.
29. Braicu OL, Budisan L, Buiga R, Jurj A, Achimas-Cadariu P, Pop LA, et al. miRNA expression profiling in formalin-fixed paraffin-embedded endometriosis and ovarian cancer samples. Onco Targets Ther. 2017;10:4225-38.
30. He J, Chang W, Feng C, Cui M, Xu T. Endometriosis Malignant Transformation: Epigenetics as a Probable Mechanism in Ovarian Tumorigenesis. Int J Genomics. 2018;2018:1465348.
31. Bastu E, Onder S, Demiral I, Ozsurmeli M, Keskin G, Takmaz O, et al. Distinguishing the progression of an endometrioma: Benign or malignant? European Journal of Obstetrics and Gynecology. 2018;230:79-84.
32. Cybulska P, Paula ADC, Tseng J, Leitao MM, Bashashati A, Huntsman DG, et al. Molecular profiling and molecular classification of endometrioid ovarian carcinomas. Gynecol Oncol. 2019;154(3):516-23.
33. Fridley BL, Dai J, Raghavan R, Li Q, Winham SJ, Hou X, et al. Transcriptomic Characterization of Endometrioid, Clear Cell, and High-Grade Serous Epithelial Ovarian Carcinoma. Cancer Epidemiol Biomarkers Prev. 2018;27(9):1101-9.
34. Tassi RA, Todeschini P, Siegel ER, Calza S, Cappella P, Ardighieri L, et al. FOXM1 expression is significantly associated with chemotherapy resistance and adverse prognosis in non-serous epithelial ovarian cancer patients. J Exp Clin Cancer Res. 2017;36(1):63.
35. Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, et al. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14(11):4376-86.
36. Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62(16):4722-9.
37. Corona RI, Seo JH, Lin X, Hazelett DJ, Reddy J, Fonseca MAS, et al. Non-coding somatic mutations converge on the PAX8 pathway in ovarian cancer. Nat Commun. 2020;11(1):2020.
38. Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature. 2020;578(7793):102-11.
39. Raphael BJ, Dobson JR, Oesper L, Vandin F. Identifying driver mutations in sequenced cancer genomes: computational approaches to enable precision medicine. Genome Med. 2014;6(1):5.
40. Carlevaro-Fita J, Lanzós A, Feuerbach L, Hong C, Mas-Ponte D, Pedersen JS, et al. Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. Commun Biol. 2020;3(1):56.
41. Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu Rev Biochem. 2015;84:405-33.
42. Li S, Xu Z, Sheng J. tRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes (Basel). 2018;9(5).
43. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452-63.
44. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7-21.
45. Cui D, Ma J, Liu Y, Lin K, Jiang X, Qu Y, et al. Analysis of long non-coding RNA expression profiles using RNA sequencing in ovarian endometriosis. Gene. 2018;673:140-8.
46. Wang JY, Lu AQ, Chen LJ. LncRNAs in ovarian cancer. Clin Chim Acta. 2019;490:17-27.
47. Lanzós A, Carlevaro-Fita J, Mularoni L, Reverter F, Palumbo E, Guigó R, et al. Discovery of Cancer Driver Long Noncoding RNAs across 1112 Tumour Genomes: New Candidates and Distinguishing Features. Sci Rep. 2017;7:41544.
48. Sun W, Yang Y, Xu C, Guo J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 2017;216-217:105-10.
49. Ferlita A, Battaglia R, Andronico F, Caruso S, Cianci A, Purrello M, et al. Non-Coding RNAs in Endometrial Physiopathology. Int J Mol Sci. 2018;19(7).
50. Marquez RT, Baggerly KA, Patterson AP, Liu J, Broaddus R, Frumovitz M, et al. Patterns of gene expression in different histotypes of epithelial ovarian cancer correlate with those in normal fallopian tube, endometrium, and colon. Clin Cancer Res. 2005;11(17):6116-26.
51. Li Q, Zhang C, Chen R, Xiong H, Qiu F, Liu S, et al. Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett. 2016;383(1):28-40.
52. Li Z, Wei D, Yang C, Sun H, Lu T, Kuang D. Overexpression of long noncoding RNA, NEAT1 promotes cell proliferation, invasion and migration in endometrial endometrioid adenocarcinoma. Biomed Pharmacother. 2016;84:244-51.
53. Smolle MA, Bullock MD, Ling H, Pichler M, Haybaeck J. Long Non-Coding RNAs in Endometrial Carcinoma. Int J Mol Sci. 2015;16(11):26463-72.
54. Zhong Y, Gao D, He S, Shuai C, Peng S. Dysregulated Expression of Long Noncoding RNAs in Ovarian Cancer. Int J Gynecol Cancer. 2016;26(9):1564-70.
55. Zhu H, Jin YM, Lyu XM, Fan LM, Wu F. Long noncoding RNA H19 regulates HIF-1?/AXL signaling through inhibiting miR-20b-5p in endometrial cancer. Cell Cycle. 2019;18(19):2454-64.
56. Hosseini E, Matthieu M, Sabzalipoor H, Kashani H, Nikzad H, Asemi Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. - PubMed - NCBI. Molecular Cancer. 2017;16(107).
57. Chen H, Tian X, Luan Y, Lu H. Downregulated Long Noncoding RNA DGCR5 Acts as a New Promising Biomarker for the Diagnosis and Prognosis of Ovarian Cancer. Technol Cancer Res Treat. 2019;18:1533033819896809.
58. Han L, Zhang W, Zhang B, Zhan L. Long non-coding RNA SOX2OT promotes cell proliferation and motility in human ovarian cancer. Exp Ther Med. 2018;15(2):2182-8.
59. Engqvist H, Parris TZ, Rönnerman EW, Söderberg EMV, Biermann J, Mateoiu C, et al. Transcriptomic and genomic profiling of early-stage ovarian carcinomas associated with histotype and overall survival. Oncotarget. 2018;9(80):35162-80.
60. Worku T, Bhattarai D, Ayers D, Wang K, Wang C, Rehman ZU, et al. Long Non-Coding RNAs: the New Horizon of Gene Regulation in Ovarian Cancer. Cell Physiol Biochem. 2017;44(3):948-66.
61. Lin X, Spindler TJ, de Souza Fonseca MA, Corona RI, Seo JH, Dezem FS, et al. Super-Enhancer-Associated LncRNA UCA1 Interacts Directly with AMOT to Activate YAP Target Genes in Epithelial Ovarian Cancer. iScience. 2019;17:242-55.
62. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991-5.
63. Wingett SW, Andrews S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res. 2018;7:1338.
64. Korpelainen Ea, Tuimala Ja, Somervuo Pa, Huss Ma, Wong Ga. RNA-seq data analysis : a practical approach. 1st ed.
65. Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, et al. The Ensembl gene annotation system. Database (Oxford). 2016;2016.
66. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760-74.
67. Mukherjee D, Fritz DT, Kilpatrick WJ, Gao M, Wilusz J. Analysis of RNA exonucleolytic activities in cellular extracts. Methods Mol Biol. 2004;257:193-212.
68. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145-66.
69. Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet. 2020;21(2):102-17.
70. Ma W, Liu Y, Ma H, Ren Z, Yan J. Cis-acting: A pattern of lncRNAs for gene regulation in induced pluripotent stem cells from patients with Down syndrome determined by integrative analysis of lncRNA and mRNA profiling data. Exp Ther Med. 2021;22(1):701.
71. Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, et al. GeneCards Version 3: the human gene integrator. Database (Oxford). 2010;2010:baq020.
72. Barshir R, Fishilevich S, Iny-Stein T, Zelig O, Mazor Y, Guan-Golan Y, et al. GeneCaRNA: A Comprehensive Gene-centric Database of Human Non-coding RNAs in the GeneCards Suite. J Mol Biol. 2021;433(11):166913.
73. RNACentral [Internet]. 2020.
74. Gong J, Shao D, Xu K, Lu Z, Lu ZJ, Yang YT, et al. RISE: a database of RNA interactome from sequencing experiments. Nucleic Acids Res. 2018;46(D1):D194-D201.
75. Karagkouni D, Paraskevopoulou MD, Tastsoglou S, Skoufos G, Karavangeli A, Pierros V, et al. DIANA-LncBase v3: indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2020;48(D1):D101-D10.
76. Ma L, Cao J, Liu L, Du Q, Li Z, Zou D, et al. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2019;47(5):2699.
77. Fang S, Zhang L, Guo J, Niu Y, Wu Y, Li H, et al. NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018;46(D1):D308-d14.
78. Landrum MJ, Chitipiralla S, Brown GR, Chen C, Gu B, Hart J, et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020;48(D1):D835-D44.
79. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353-D61.
80. Belinky F, Nativ N, Stelzer G, Zimmerman S, Iny Stein T, Safran M, et al. PathCards: multi-source consolidation of human biological pathways. Database (Oxford). 2015;2015.
81. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-d13.
82. Cui D, Liu Y, Ma J, Lin K, Xu K, Lin J. Identification of key genes and pathways in endometriosis by integrated expression profiles analysis. PeerJ. 2020;8:e10171.
83. Zheng J, Zhou Z, Qiu Y, Wang M, Yu H, Wu Z, et al. A Prognostic Ferroptosis-Related lncRNAs Signature Associated With Immune Landscape and Radiotherapy Response in Glioma. Front Cell Dev Biol. 2021;9:675555.
84. Luo Y, Huang Q, He B, Liu Y, Huang S, Xiao J. Regulation of ferroptosis by non?coding RNAs in the development and treatment of cancer (Review). Oncol Rep. 2021;45(1):29-48.
85. Jenkins T, Gouge J. Nrf2 in Cancer, Detoxifying Enzymes and Cell Death Programs. Antioxidants (Basel). 2021;10(7).
86. Kang YP, Mockabee-Macias A, Jiang C, Falzone A, Prieto-Farigua N, Stone E, et al. Non-canonical Glutamate-Cysteine Ligase Activity Protects against Ferroptosis. Cell Metab. 2021;33(1):174-89.e7.
87. Deng HF, Yue LX, Wang NN, Zhou YQ, Zhou W, Liu X, et al. Mitochondrial Iron Overload-Mediated Inhibition of Nrf2-HO-1/GPX4 Assisted ALI-Induced Nephrotoxicity. Front Pharmacol. 2020;11:624529.
88. Yuan Y, Zhai Y, Chen J, Xu X, Wang H. Kaempferol Ameliorates Oxygen-Glucose Deprivation/Reoxygenation-Induced Neuronal Ferroptosis by Activating Nrf2/SLC7A11/GPX4 Axis. Biomolecules. 2021;11(7).
89. Ng A, Ng S, Arcuri F, Toti P, Genega E, Norwitz E, et al. Iron Overload and Ferroptosis in Endometriosis and Endometriosis-associated ovarian cancer. 67th Annual Scientific Meeting of the Society for Reproductive Investigation; Vancouver, Canada: Reproductive Sciences; 2020. p. 156A.
90. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339-46.
91. Gao N, Li Y, Li J, Gao Z, Yang Z, Liu H, et al. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front Oncol. 2020;10:598817.
92. Akrami R, Jacobsen A, Hoell J, Schultz N, Sander C, Larsson E. Comprehensive analysis of long non-coding RNAs in ovarian cancer reveals global patterns and targeted DNA amplification. PLoS One. 2013;8(11):e80306.
93. Sang B, Zhang YY, Guo ST, Kong LF, Cheng Q, Liu GZ, et al. Dual functions for OVAAL in initiation of RAF/MEK/ERK prosurvival signals and evasion of p27-mediated cellular senescence. Proc Natl Acad Sci U S A. 2018;115(50):E11661-E70.
94. Zou AE, Ku J, Honda TK, Yu V, Kuo SZ, Zheng H, et al. Transcriptome sequencing uncovers novel long noncoding and small nucleolar RNAs dysregulated in head and neck squamous cell carcinoma. RNA. 2015;21(6):1122-34.
95. Azmi AS, Bao GW, Gao J, Mohammad RM, Sarkar FH. Network insights into the genes regulated by hepatocyte nuclear factor 4 in response to drug induced perturbations: a review. Curr Drug Discov Technol. 2013;10(2):147-54.
96. Kapoor RR, Locke J, Colclough K, Wales J, Conn JJ, Hattersley AT, et al. Persistent hyperinsulinemic hypoglycemia and maturity-onset diabetes of the young due to heterozygous HNF4A mutations. Diabetes. 2008;57(6):1659-63.
97. Fadare O, Liang SX. Diagnostic utility of hepatocyte nuclear factor 1-beta immunoreactivity in endometrial carcinomas: lack of specificity for endometrial clear cell carcinoma. Appl Immunohistochem Mol Morphol. 2012;20(6):580-7.
98. Drewes T, Senkel S, Holewa B, Ryffel GU. Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol Cell Biol. 1996;16(3):925-31.
99. Allen WE, Zicha D, Ridley AJ, Jones GE. A role for Cdc42 in macrophage chemotaxis. J Cell Biol. 1998;141(5):1147-57.
100. Howe KL, Achuthan P, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, et al. Ensembl 2021. Nucleic Acids Res. 2021;49(D1):D884-D91.
101. Wu Y, Zhao Y, Yang HZ, Wang YJ, Chen Y. HMGB1 regulates ferroptosis through Nrf2 pathway in mesangial cells in response to high glucose. Biosci Rep. 2021;41(2).
102. Lei P, Bai T, Sun Y. Mechanisms of Ferroptosis and Relations With Regulated Cell Death: A Review. Front Physiol. 2019;10:139.
103. Ng SW, Norwitz SG, Taylor HS, Norwitz ER. Endometriosis: The Role of Iron Overload and Ferroptosis. Reprod Sci. 2020;27(7):1383-90.
104. Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266-82.
105. Stockwell BR, Jiang X, Gu W. Emerging Mechanisms and Disease Relevance of Ferroptosis. Trends Cell Biol. 2020;30(6):478-90.
106. Luzón-Toro B, Fernández RM, Martos-Martínez JM, Rubio-Manzanares-Dorado M, Antiñolo G, Borrego S. LncRNA LUCAT1 as a novel prognostic biomarker for patients with papillary thyroid cancer. Sci Rep. 2019;9(1):14374.
107. Kobayashi H, Kimura M, Maruyama S, Nagayasu M, Imanaka S. Revisiting estrogen-dependent signaling pathways in endometriosis: Potential targets for non-hormonal therapeutics. Eur J Obstet Gynecol Reprod Biol. 2021;258:103-10.
108. Shang R, Wang M, Dai B, Du J, Wang J, Liu Z, et al. Long noncoding RNA SLC2A1-AS1 regulates aerobic glycolysis and progression in hepatocellular carcinoma via inhibiting the STAT3/FOXM1/GLUT1 pathway. Mol Oncol. 2020;14(6):1381-96.
109. Hayashi M, Sakata M, Takeda T, Yamamoto T, Okamoto Y, Sawada K, et al. Induction of glucose transporter 1 expression through hypoxia-inducible factor 1alpha under hypoxic conditions in trophoblast-derived cells. J Endocrinol. 2004;183(1):145-54.
110. Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171(2):273-85.
111. Yu H, Han Z, Xu Z, An C, Xu L, Xin H. RNA sequencing uncovers the key long non-coding RNAs and potential molecular mechanism contributing to XAV939-mediated inhibition of non-small cell lung cancer. Oncol Lett. 2019;17(6):4994-5004.
112. Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23(6):768-83.
113. Russell MR, Penikis A, Oldridge DA, Alvarez-Dominguez JR, McDaniel L, Diamond M, et al. CASC15-S Is a Tumor Suppressor lncRNA at the 6p22 Neuroblastoma Susceptibility Locus. Cancer Res. 2015;75(15):3155-66.
114. Valero R, Marfany G, González-Angulo O, González-González G, Puelles L, Gonzàlez-Duarte R. USP25, a novel gene encoding a deubiquitinating enzyme, is located in the gene-poor region 21q11.2. Genomics. 1999;62(3):395-405.
115. Zhong B, Liu X, Wang X, Li H, Darnay BG, Lin X, et al. Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitination of the adaptor protein TRAF3. Sci Signal. 2013;6(275):ra35.
116. Yang L, Chen X, Yang Q, Chen J, Huang Q, Yao L, et al. Broad Spectrum Deubiquitinase Inhibition Induces Both Apoptosis and Ferroptosis in Cancer Cells. Front Oncol. 2020;10:949.
117. Zhang X, Wang L, Li H, Zhang L, Zheng X, Cheng W. Crosstalk between noncoding RNAs and ferroptosis: new dawn for overcoming cancer progression. Cell Death Dis. 2020;11(7):580.