Abstract
Cancer mortality is proportionally higher in Africa than elsewhere in the world. In Senegal, ovarian cancer is responsible for 2.8% of deaths and is one of the most fatal gynaecological cancers. This work is therefore being carried out in order to better understand the impact of D-Loop mutations in the evolution of ovarian cancer in Senegalese women. The genetic variability of D-Loop was studied by PCR-Sequencing, in thirty-eight (38) patients. The results revealed twenty-two (22) mutations. In this study, eleven (11) mutations (A66G, C186T, G309A, C388T, C418T, G421T, C422A, A425C, A425G, C431T and G499A) were identified as somatic mutations in ovarian cancer. All the variants are present in the Mitomap database. A variation (19.05%) in the length of the first section of the D310 area was found. The results obtained in this study show that the D310 region of the D-Loop is not the “hot spot” for mtDNA mutations in ovarian cancer in Senegalese women.
Keywords
Cancer, Ovary, Displacement Loop, Mutations
Introduction
Each cell in the body contains hundreds of mitochondria; each mitochondrion contains several DNA molecules, each carrying 37 genes that participate in the production of energy. The mitochondria are organelles that function in a semi-autonomous manner, containing their own genome and are capable of replication and transcription. They are responsible for more than 90% of cellular ATP production through the process of oxidative phosphorylation [1]. Studies on mitochondria have shown that alterations in mtDNA are closely linked to hereditary diseases, ageing and cancer [2]. Indeed, the involvement of mitochondria in apoptosis and probably in tumorigenesis has raised interest in the potential role of mtDNA mutations in the development of cancers [3].
Despite the small size of the mitochondrial genome, mtDNA mutations are an important cause of hereditary diseases. Because mitochondria lack protective histones or highly efficient DNA repair mechanisms, mammalian mtDNA accumulates at a rate 10 times higher than nuclear DNA [4]. In recent years, considerable progress has been made in understanding mitochondrial genetics and identifying mutations in mtDNA acquired in cancers [5].
For example, genetic mapping and cloning of chromosomal “oncogenes” has revealed that some cancers are caused by mutations in genes of nucleusencoded mitochondrial enzymes [6]. These alterations are particularly frequent in pre-neoplastic lesions and in human cancers, particularly in breast, ovarian, colon and rectal, stomach, liver and oesophagus cancers [7].
Cancers are among the leading causes of morbidity and mortality worldwide. More than 60% of new cancer cases occur in Africa, Asia, Central and Latin America. Cancer mortality is proportionally higher in Africa than elsewhere in the world [8]. Cancers already account for 10-20% of pathologies on the African continent [8]. Thus, ovarian cancers are among the gynecological cancers of women and have a high mortality and morbidity rate worldwide and particularly in Africa [9]. In Senegal, 260 new cases of ovarian cancer have been detected and the mortality rate has been estimated at 2.8% [10].
In addition to the degenerative diseases that dominate the health picture in our societies, cancer, or rather cancers, remains one of the most complex challenges facing medicine in terms of understanding these pathologies, diagnosis and treatment. It is in this sense that this study aims to understand the impact of D-Loop mutations in the evolution of ovarian cancers.
Materials and Methods
Subjects
The study was approved by the University Cheikh Anta Diop Research Ethics committee. Informed consent was obtained from all subjects prior to enrollment. The tissues samples were derived from thirty-eight (38) patients. These tissues were obtained following surgery performed at the cancerology department of the Joliot Curie Institute of the Aristide Le Dantec Hospital in Dakar. They were preserved in 96% alcohol. Blood samples were taken from healthy control subjects (N=10).
DNA isolation, amplification and sequencing
The extraction of DNA from the tissue and blood was performed using the Kit Qiagen Dneasy method. The PCR required a reaction volume of 50 μl containing 34.9 μl milliQ water, 5 μl buffer 10X, 2 μl dNTP, 2.5 μl for each primer (H408 and L16340), 0.1 μl Taq polymerase and 2 μl cDNA. Briefly, total DNA was subjected to the following PCR protocol: initial DNA denaturization at 95°C for 15 min, followed by 35 cycles at 95°C for 30 s, 62°C for 30 s, 72°C for 2 min, and a final extension at 72°C for 10 min. Positive PCR products were purified and sequenced. Sequencing reactions were performed in an MJ Research PTC-225 Peltier thermal cycler with the ABIPRISM Kit and subjected to electrophoresis in the ABI3730 XL sequencer.
Identification of somatic mutations
The resulting sequences were cleaned, corrected and aligned with Bioedit software version 7.1.9 [11] using the ClustalW algorithm [12]. Alignment is indeed an important step in the data analysis, as it determines the homology of the sites. The sequences of the diseased individuals were compared with those of the controls in order to identify the different variants of D-Loop in ovarian cancer. Any variant that is present in the controls is considered a polymorphism. The variant is considered a mutation if it is present only in patients. We consulted the mitochondrial database MITOMAP (www.mitomap. org) which provides users with detailed information on sequence, function, polymorphisms and mutations in germ and somatic lines associated with disease states [13]. Variants not found in the database have been recorded as new mutations in ovarian cancer. This test is used to verify the difference in the probability of mutation in populations with and without cancer [14]. To highlight the polymorphism of the D310 region, we performed a comparison of this region between controls and patients. Significance testing is performed by the chi2 test with Statview software version 5.0 (Roach, 2004). For all tests performed, the level of significance is 5%.
Polymorphism and genetic diversity
For this analysis, two groups were formed: healthy tissue and cancerous tissue. For each group, all the basic parameters of genetic variability: number of sites (N), sample size (n), number of variable sites, number of haplotypes (h), mean number of nucleotide differences (k) between sequences were estimated using DnaSP version 5.10 software [15]. However, the nucleotide frequencies of D-Loop in controls and patients, the nature of mutations (percentage of transitions and transversions), and the mutation rate (R) were calculated using MEGA7 version 7.0.14 [16].
Genetic differentiation
The estimation of genetic differentiation between control and patient sequences generally requires two indices, genetic distance (D) and FST. Genetic distance is determined within each population (intra-population genetic distance) and between populations taken in pairs (inter-population genetic distance). Inter-population TST values were evaluated using the Harlequin program version 3.5.1.3 [17]. Intra- and inter-population genetic distances were derived using MEGA7 version 7.0.14 [16].
Neutrality tests
In order to study the genetic evolution of tumour D-Loop mutations, the indices of haplotypic diversity (hd) and nucleotide diversity (Pi) [18] and their variance were calculated with DnaSP version 5.10 [15]. The indices of the D of Tajima [19], the D* and F* of Fu and Li, and the Fs of Fu [20] were used to test the deviation from the neutrality hypothesis using Arlequin software version 3.5.1.3 [17] and with DnaSP version 5.10 [15]. The Mismatch analysis combines two indices, SSD (sum of deviation squares) and Rag (irregularity index), which test the goodness of fit of the distribution. The graphs are constructed using DnaSP version 5.10 [15]. The SSD and Rag indices are estimated using Arlequin software version 3.5.1.3 [17]. The purpose of these tests is to distinguish sequences, whose evolution follows a neutral evolutionary model, and those evolving according to a non-random process.
Results
D-Loop somatic mutations
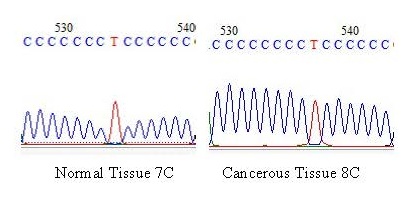
Table 1 shows the positions and frequencies of the D-Loop mutations. We first verified the reliability of the sequence using the Nucleotide Blast algorithm with the Cambridge reference sequence under accession number (NC_012920) in order to list the mutation positions at the mitochondrial genome level in the Mitomap database. The portion of DNA studied extended from position 16390 to position 358 and the majority of mutations were found in the hypervariable zone (HV2) and only the A66G and C186T mutations were located on the MT-ATT locus. Analysis of the nucleotide sequences revealed twenty-six (26) variations. However, the variant is only considered a mutation when it is present only in cancerous tissues, therefore twenty-two (22) mutations were detected with more transitions (18) than transversions (4). The A57G variant is the only one present in controls with a significant P value (0.003). In this study, eleven (11) mutations (A66G, C186T, G309A, C388T, C418T, G421T, C422A, A425C, A425G, C431T and G499A) out of twenty-two (22) or 50% of the variants were identified as somatic mutations in ovarian cancer, no new mutation was found because all the variants are present in the Mitomap database. All variants show significant differences between controls and cancerous tissues with a P value less than 0.05 except for the C186T and C418T mutations which are non-significant with a P value greater than 0.05. In cancerous tissues, there are four individuals (19.05%) with cytosine elongation in the D310 region, thus having eight (8) cytosines instead of seven (7) as observed in controls (Figure 1).
| Mutations | Type | Controls (%) |
TC (%) |
rRCS | Locus | Mitomap | Associated Diseases | Ref | P-value |
|---|---|---|---|---|---|---|---|---|---|
| A57G | Ts | 10 | 0 | 16390 | ATT | + | Breast cancer. ovarian tumour | Ref | 0.003* |
| A66G | Ts | 0 | 4.76 | 16399 | ATT | + | Gastrique carcinome | Ref | 0.0015* |
| C186T | Ts | 0 | 38.09 | 16519 | ATT | + | Ovarian tumors. prostate | Ref | 0.102 |
| G309A | Ts | 0 | 4.76 | 73 | HV2 | + | Thyroid tumors. prostate | Ref | 0.0015* |
| T325C | Ts | 0 | 4.76 | 89 | HV2 | + | - | - | 0.0015* |
| A329G | Ts | 10 | 14.38 | 93 | HV2 | + | - | - | - |
| A331C | Tv | 0 | 4.76 | 95 | HV2 | + | - | Ref | 0.0015* |
| G379A | Ts | 0 | 4.76 | 143 | HV2 | + | - | - | 0.0015* |
| C382T | Ts | 10 | 66.67 | 146 | HV2 | + | Ovarian Tumor | Ref | - |
| C386T | Ts | 10 | 42.86 | 150 | HV2 | + | Thyroid tumors. lung tumors | Ref | - |
| C387T | Ts | 0 | 4.76 | 151 | HV2 | + | - | - | 0.0015* |
| C388T | Ts | 0 | 23.81 | 152 | HV2 | + | Ovarian tumor. breast tumor | - | 0.006* |
| C418T | Ts | 0 | 38.09 | 182 | HV2 | + | - | Ref | 0.102 |
| G421T | Tv | 0 | 9.52 | 185 | HV2 | + | - | Ref | 0.002* |
| C422A | Tv | 0 | 4.76 | 186 | HV2 | + | - | Ref | 0.0015* |
| A425C | Tv | 0 | 9.52 | 189 | HV2 | + | - | Ref | 0.002* |
| A425G | Ts | 0 | 4.76 | 189 | HV2 | + | - | Ref | 0.0015* |
| C431T | Ts | 0 | 33.33 | 195 | HV2 | + | Tumors of the lung. ovary | Ref | 0.009* |
| C434T | Ts | 0 | 28.57 | 198 | HV2 | + | - | - | 0.008* |
| T440C | Ts | 0 | 14.28 | 204 | HV2 | + | - | Ref | 0.004* |
| G443A | Ts | 0 | 4.76 | 207 | HV2 | + | Oral tumors. thyroid tumors | Ref | 0.0015* |
| G483A | Ts | 0 | 14.28 | 247 | HV2 | + | - | - | 0.004* |
| G499A | Ts | 0 | 4.76 | 263 | HV2 | + | - | Ref | 0.0015* |
| G554A | Ts | 0 | 4.76 | 317 | HV2 | + | Ovarian Tumor | Ref | 0.0015* |
| C563T | Ts | 0 | 19.05 | 326 | HV2 | + | - | - | 0.005* |
| A595G | Ts | 0 | 9.52 | 358 | HV2 | + | - | - | 0.002* |
| +: indicates that the mutation is present in Mitomap, -: indicates that the mutation was not found in Mitomap, * : indicates a significant value, Ref: indicates that the variant is referenced, Ts: Transition, Tv: Transversion | |||||||||
Genetic diversity of D-Loop
Table 2 shows the different parameters of the genetic diversity of controls and cancerous tissues. It was noted that the number of variable sites is higher in cancer tissues (23) compared to controls (4). Nucleotide frequencies show that C and A bases are more frequent with 30.2% and 27.6% for controls and 29.8% and 27.6% for cancerous tissues respectively. Transitions are higher in controls (99.98%) than in cancerous tissues (87.51%). No transversion was noted in the controls in contrast to the cancerous tissues with 12.48% of transversions and the ratio of transitions to transversions was estimated to be 7.01.
| Controls | Cancerous tissues | |||||||
|---|---|---|---|---|---|---|---|---|
| Number of sites | 662 | 662 | ||||||
| Number of sequences | 10 | 21 | ||||||
| Variable number of sites | 4 | 23 | ||||||
| Number of haplotypes | 2 | 15 | ||||||
| Mean number of nucleotide differences | 0.800 | 5.829 | ||||||
| Nucleotide frequencies | T | C | A | G | T | C | A | G |
| 25.4 | 30.2 | 27.6 | 16.8 | 25.8 | 29.8 | 27.6 | 16.8 | |
| Transitions (%) | 99.98 | 87.51 | ||||||
| Transversions (%) | 00 | 12.48 | ||||||
| R(Ts/Tv) | - | 7.01 | ||||||
Genetic Differentiation of D-Loop
The results show that the value of the genetic distance between cancerous tissues (0.009) is greater than that observed between controls (0.001). The genetic distance between controls and cancerous tissues is 0.007 closer to that observed between cancerous tissues. The degree of genetic differentiation (0.219) is significant (P=0.023) Table 3.
| Within-population genetic distances | Between-population genetic distances |
Fst | |
|---|---|---|---|
| Controls | 0,001 | 0,007 | 0,219 P value=0,023 |
| Cancerous tissues | 0,009 |
Genetic evolution of D-Loop
Table 4 shows a high haplotypic diversity (Hd=0.922) and a low nucleotide diversity (Pi=0.017) for cancerous tissues. The statistical values of Tajima’s D (-0.064) and Fu and Li D* (-0.041) appear negative and nonsignificant while those of FU Fs (0.305) and F* Fu and Li (0.173) are positive and also non-significant with P values greater than 0.05.
| Hd | Pi | Tajima D | FU Fs | D* | F* |
|---|---|---|---|---|---|
| 0,922 | 0,017 ± 0,008 | -0,064 ±0,552 | 0,305 ± 0,463 | -0,041 ± 0,462 | 0,173 ±0,506 |
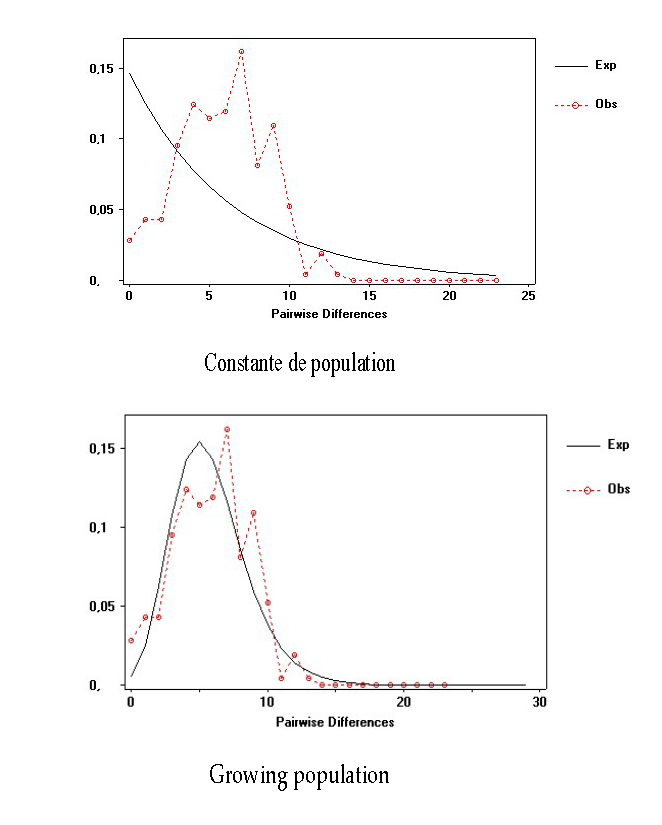
Graphical representations of the genetic distances between cancerous tissues of individuals taken in pairs, assuming a constant or expanding population, show a multimodal distribution. The SSD (0.0025; P=0.072) and Rag (0.045; P=1.30) indices with non-significant P values show a growing population at the cancer tissue level.
Figure 2: Mismatch distribution curves of D-Loop for cancerous tissues.
Discussion
Our study focuses on D-Loop, which is the non-coding and regulatory region of the mitochondrial genome. Its general objective is to determine the impact of D-Loop mutations on the evolution of ovarian cancers. The control region of the mtDNA is highly polymorphic and contains the hypervariable HV2 zone characterized by a high degree of polymorphism [21]. It is in the latter that most of the variants (88.46%) found in this study is located. 81.82% and 18.18% of the mutations are transitions and transverse respectively. This is in line with the assertion of Li and Hong [22], who showed in their study that the majority of cancer cell mutations are transition-type base substitutions. The A57G variant is the only one present in controls with a significant P value (0.003). It could therefore have a protective action against ovarian tumor as well as the C186T and C418T mutations present in TCs but not significantly. Mutations (A66G, G309A, T325C, A331C, G379A, C387T, C388T, G421T, C422A, A425C, A425G, C431T, C434T, T440C, G443A, G483A, G499A, G554A, C563T and A595G) are present only in cancerous tissues with significant P values. They may play an important role in the development of ovarian tumors. In this study eleven (11) mutations (A66G, C186T, G309A, C388T, C418T, G421T, C422A, A425C, A425G, C431T and G499A) out of twenty-two (22) or 50% of the variants were identified in Mitomap as somatic mutations in ovarian cancer. These results are in agreement with those of Bragoszewski et al. [7] who showed in their study the presence of somatic D-Loop mutations in 50% of ovarian cancer tissue samples, and most of the samples carried a single mutation. These results are consistent with previous reports describing somatic D-Loop mutations in 20-78% of human cancers. Most of the somatic mtDNA mutations identified in human cancers (bladder, head and neck, lung, breast, ovary and esophagus) are found in the hypervariable region of the promoter. Generally, it is concluded that the majority of the variants found are actually acquired somatically. According to Chen et al. [23], assuming that the polymorphic sites of D-loop have neutral function, these somatic mutations could confer functional consequences on cancer cells. Therefore, this also implies a possible impact of the genetic structure of D-Loop on the mode of mutagenesis of mtDNA in human tumors.
The polymorphism of the D310 region was also studied in this study. The microsatellite (D310) located between positions 303-315 of the D-Loop is used as a potential marker for early cancer detection [24]. 19.05% of sick individuals have a cytosine insertion, therefore, we can hypothesize that cytosine elongation could be a risk factor for developing ovarian cancer. Indeed, the hot spot for instability has a unique sequence pattern between nucleotides 303 and 316 (CCCCCCCTCCCCCC), and instability has always occurred in the first polymorphic C tract, previously called D310, which is particularly rich in somatic mutations in several types of cancer [1]. It should be noted that in a previous study, this mutation hot spot was considered a major target for mtDNA alterations in human tumors [23]. The occurrence of mutations in this single nucleotide sequence could therefore alter this regulatory function and increase the replication rate of the mtDNA, thus generating a replicative advantage at the level of the mutated mitochondria that is responsible for an accumulation of mutations during successive divisions [1].
Parameters such as the basic parameters of genetic diversity have also been studied showing tumor variation. Indeed, tumor cells are generally characterized by faster cell proliferation, due to the presence of alterations in the genes that regulate proliferation in normal cells [25]. It is in fact mutations that are the ultimate source of genetic variation that cause diversity in all organisms, thus playing the driving role in evolution [25]. However, according to Moto Kimura’s model known as the theory of mutation and random drift, most genetic variability is neutral and polymorphisms are eliminated or fixed in individuals under the influence of the effects of genetic drift from the environment.
Analysis of the genetic differentiation of D-Loop revealed that the genetic distance between cancerous tissues (0.009) is greater than that observed between controls (0.001). The proliferation of cancer cells appears to be much faster, also indicating tumor homogeneity. In contrast, the inter-population genetic distance (0.007) between controls and cancerous tissues is closer to that of cancerous tissues. The degree of genetic differentiation (Fst) has a significant P value (0.023) indicating genetic differentiation between groups.
The study of the genetic evolution of D-Loop mutations in ovarian cancer in Senegalese women is marked by high polymorphism with high haplotypic diversity (Hd=0.922) and low nucleotide diversity (Pi=0.017) in cancerous tissues. D-Loop mutations are therefore believed to grow rapidly in ovarian tumors. Cancer cells contain many clonal mutations, i.e. mutations that are present in most or all malignant cells in a tumor and they confer a proliferation advantage. Thus, a malignant tumor is a disease characterized by abnormally high cell proliferation within normal body tissue, such that the survival of the body is threatened. Tests of the neutrality of cancerous tissues have shown that the D of tajima and the D* of Fu and Li are negative and insignificant, which makes it possible to affirm that the mutations found are neutral. Indeed, for non-coding sequences such as the mtDNA control region, a discrepancy in neutrality is probably explained by recent demographic changes rather than by selection [25]. Moderate population growth in cancerous tissues is also confirmed by the multimodal distribution of mismatch curves.
Conclusion
Ovarian cancer is a morphologically and biologically heterogeneous disease which has probably contributed to the difficulty in defining the molecular alterations associated with its development and progression. It is the second most common form of cancer in the female reproductive system and the most lethal of the gynecological malignancies. For decades, considerable efforts have been made to shed light on the molecular basis for the initiation and progression of ovarian carcinoma. Our objective was to evaluate the involvement of D-Loop mutations in ovarian cancers in Senegalese women. Our study revealed a strong polymorphism in ovarian cancer in Senegalese women. The results revealed mutations with significant P values, which could be implicated in an increased risk and play an important role in the evolution of ovarian carcinomas. With the exception of the C186T and C418T mutations present in cancerous tissues, but not significantly, they could therefore have a protective action against the ovarian tumor. In addition, the study of genetic diversity parameters revealed a variation in tumors. The results also showed a genetic difference between controls and cancerous tissues and a much more rapid proliferation can be noted in the cancerous tissues.
References
2. Van Trappen PO, Cullup T, Troke R, Swann D, Shepherd JH, Jacobs IJ, et al. Somatic mitochondrial DNA mutations in primary and metastatic ovarian cancer. Gynecologic Oncology. 2007 Jan 1;104(1):129-33.
3. Liu VW, Shi HH, Cheung AN, Chiu PM, Leung TW, Nagley P, et al. High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas. Cancer Research. 2001 Aug 15;61(16):5998-6001.
4. Yoneyama H, Hara T, Kato Y, Yamori T, Matsuura ET, Koike K. Nucleotide sequence variation is frequent in the mitochondrial DNA displacement loop region of individual human tumor cells. Molecular Cancer Research. 2005 Jan 1;3(1):14-20.
5. Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nature Reviews Genetics.2005 May;6(5):389-402.
6. Brandon M, Baldi PA, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006 Aug;25(34):4647-62.
7. Bragoszewski P, Kupryjanczyk J, Bartnik E, Rachinger A, Ostrowski J. Limited clinical relevance of mitochondrial DNA mutation and gene expression analyses in ovarian cancer. BMC Cancer. 2008 Dec 1;8(1):292.
8. GLOBOCAN. 2017. Les cancers en Afrique francophone- ICCP Portal. https://www.iccp-portal.org.Accédé le 4 avril 2019.
9. Sando Z, Mboudou E, Fouogue TJ, Nganwa G, Tchuendem J, Essame JL, Doh AS, Genton CY. Clinical and pathological profile of ovarian cancers in Yaounde-Cameroun. Clinics in Mother and Child Health. 2010;7(1):1183-7.
10. GLOBOCAN. 2018. The Global Cancer Observatory. Accédé le 6 décembre 2019.
11. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. InNucleic acids symposium series 1999 Jan 1 (Vol. 41, No. 41, pp. 95-98). [London]: Information Retrieval Ltd., c1979-c2000.
12. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994 Nov 11;22(22):4673-80.
13. Cohen EE. A Disturbance in the Force—Mitochondrial Mutations in Squamous Cell Carcinoma of the Head and Neck. Clinical Cancer Research. 2007 Aug 1;13(15):4317-9.
14. Sultana GN, Rahman A, Shahinuzzaman AD, Begum RA, Hossain CF. Mitochondrial DNA mutations—candidate biomarkers for breast cancer diagnosis in Bangladesh. Chinese Journal of Cancer. 2012 Sep;31(9):449.
15. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data.Bioinformatics. 2009 Jun 1;25(11):1451-2.
16. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016 Mar 22;33(7):1870-4.
17. Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology resources. 2010 May;10(3):564-7.
18. Nei M. Molecular Evolutionary Genetics. Columbia University Press. New York, 1987.
19. Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989 Nov 1;123(3):585-95.
20. Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997 Oct 1;147(2):915-25.
21. Calloway CD, Reynolds RL, Herrin Jr GL, Anderson WW. The frequency of heteroplasmy in the HVII region of mtDNA differs across tissue types and increases with age. The American Journal of Human Genetics. 2000 Apr 1;66(4):1384-97.
22. Li H, Hong ZH. Mitochondrial DNA mutations in human tumor cells. Oncology Letters. 2012 Nov 1;4(5):868-72.
23. Chen JZ, Gokden N, Greene GF, Mukunyadzi P, Kadlubar FF. Extensive somatic mitochondrial mutations in primary prostate cancer using laser capture microdissection. Cancer Research. 2002 Nov 15;62(22):6470-4.
24. Troudi W, Loueslati B, Baccar A, Ben FA, Ben AA. Penetrance of BRCA1 gene mutation and DNA mitochondrial in Tunisian breast cancer occurrence. La Tunisie Medicale. 2009 Aug;87(8):494-8.
25. Doupa D. Benign Breast Tumors among Senegalese Women: Diversity and Genetic Evolution of D-Loop.Open Access Library Journal. 2015;2(08):1.