Abstract
HPV is the most common sexual transmitted disease worldwide. There is no specific treatment for the disease and the impact of HPV on public health and the necessity for more prominent diagnosis and treatment are reflected by the genital infection and cervical cancer statistics. The disease has a high rate of deaths due to its relation to cancer, principally cervix cancer. Cancerous warts also affect the vulva, vagina, anus, penis, mouth and throat. There are more than 80 HPV types and more than 40 infect the genital tract. Several HPV genes and proteins are potential candidates as genetic markers for the development of cancer and they are target for vaccines and HPV treatment. E6 exerts pleiotropic functions, including signaling, cell cycle regulation, cell line transformation, immortalization of primary cell line and genome stability regulation. The E6 protein interacts with several host-proteins, including TRAF5, which is related to cytokines signaling. E6 induces the destruction of host regulatory proteins through the proteasome system, being an important target for the development of new therapies against HPV. Here, we hypothesize that interaction between TRAF5 and E6 could be modulated in order to inhibit the activity of E6. We have shown an in silico approach of interaction between TRAF5 and HPV E6 through the identification of hot spots within the interface of interaction of the complex. We propose a new peptide that interacts and inhibits HPV E6. For a future perspective, the peptide designed will be tested in vitro and other regions of the interface of interaction will also be screened so that other peptides could also be designed and tested.
Keywords
HPV; TRAF5; hot spots screening; design of peptide
Introduction
According to the World Health Organization (WHO), the human papillomavirus (HPV) affects more than 600 million people worldwide, being the most common sexually transmitted disease (STD). There were over 250,000 deaths due to cervical cancer worldwide and most of them took place in developing countries (WHO). There are more than 80 HPV types and more than 40 infect the genital tract [1]. Chronic infection with certain HPV subtypes increases cervical cancer susceptibility [2], even though most HPV infections are asymptomatic and warts might spontaneously resolve [3]. The immune system gets rid of HPV within 12-24 months in 90% of the patients [4], either completely or reduced to undetectable levels [5]. Cancerous warts affect mainly the uterine canal and less frequently the vulva, vagina, anus, penis, mouth and throat [6,7]. There is no specific treatment for the disease and the impact of HPV on public health and the necessity for more prominent diagnosis and treatment are reflected by the genital infection and cervical cancer statistics.
HPV infects epithelial cells and it takes around 24 hours for the beginning of viral components transcription to occur, with up to 200 viral genomes in each host cell infected [8]. The immune system cells are activated but viral particles still remain on the cell surfaces, bound to receptors such as annexin, integrin and laminins [9]. The HPV genome comprises 8 Kbp (kilo base pairs) in a circular double DNA strand [10,11]. The viral genome contains two main regions. The late region (L), which codes for the capsid proteins and the early region (E) with sequences that code for proteins related to transcription and cellular transformation. In addition, genes within the E region has a central role as oncogenes and are usually selected as possible target for diagnosis and drug development [12].
It has been shown that several types of cytokines inhibit intracellular HPV proliferation and the expression of the E region genes [13]. The cytokine tumor necrosis factor (TNF) is produced by keratinocytes, which are host to HPV. These cells show anti-proliferative properties on infected cells [14,15]. TNF, transforming growth factor (TGF) and interleukin 1α repress certain types of HPV E6 and E7 oncogenes expression in epithelial cells [16]. Malignant transformation arises with the loss of the inhibitory roles of cytokines due to polymorphisms [17], family history [18], environmental [19] and xenobiotic factors [20]. Keratinocytes express receptors with innate immune system roles on their plasma membrane such as pathogen recognition receptors, thus the immune system recognize pathogen associated molecular patterns [21]. Innate and adaptive immune cells respond to viruses trying to stop the infection and to eliminate the infected cells. However, the pathogens down regulate cytokine responses in order to control cellular metabolism and establish infection.
Some of the proteins coded by the E region of the HPV genome, especially the protein E6, show cell transforming properties [22] and being requested for malignant transformation. E6 exerts pleiotropic functions, including signaling [23], cell cycle regulation [24], cell line transformation [22], immortalization of primary cell line [25] and genome stability regulation [26]. The E6 protein interacts with several host-proteins and induces the destruction of host regulatory proteins through the proteasome system [27]. The protein has 158 amino acid residues and its conformational tridimensional structure is formed by two zinc-finger binding motif, PDZ-binding domain, an endonuclease and a helicase domains [28]. The E6 oncogene might compromise the immune system through down regulation of antigen-presenting cells, induce viral persistence and increased susceptibility of cancer. Moreover, super expression of E6 and other oncogenes increases genomic instability, bringing opportunity to cancer cells scape immune surveillance.
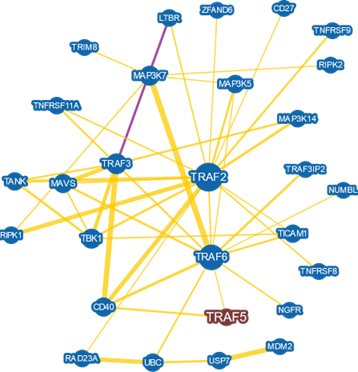
Cytokines regulates the immune response of the HPV infection. An important family of cytokines related to HPV and the viral immune evasion is the tumor necrosis factor receptor-associated factor (TRAF), as it mediates TNF activation. It has been shown that HPV E proteins increases the effect of TNF through TRAF5 activation by direct interaction [29]. In addition, TRAF5 is down regulated in cytoplasmic extracts from cells infected with HPV compared to uninfected control cells [30]. Hence, defects of genes coding for cytokines and other immune system-related proteins produce a favorable microenvironment to HPV replication, lesion endurance and immune evasion, increasing HPV patients’ susceptibility to cancer.
Here, we hypothesize that interaction between TRAF5 and E6 could be modulated in order to inhibit the activity of E6. We have shown an in silico approach of interaction between TRAF5 and HPV E6 through the identification of hot spots within the interface of interaction of the complex. We propose a new peptide that interacts and inhibits HPV E6 functions.
Materials and Methods
The 3-D structures of the Homo sapiens protein TRAF5 and the HPV E6 used in the analysis are available in the PDB (protein databank; https://www.rcsb.org/). We used KBDOCK and InterPro in order to find the E6 and TRAF5 protein domains [28,31,32]. The TRAF5 interactome was performed on BioGrid and included protein-protein interaction with evidence from genetic and physical experimental. For each interaction in the interactome was set a minimum evidence score of 5, which means that the interaction was validated by at least five different types approaches [33]. Docking analysis was performed through ClusPro [34]. The procedure is based on Piper [35], which relies on a multi-staged approach and numerical methods to predict protein-protein interactions. The models of interactions are grouped into clusters according to their size and the they are ranked. The final best model relies on the one that showed lower free binding energy.
We used PyMol (https://pymol.org) in order to analyze the docking results by the visualization of the interface of interaction, predicted hot spots and polymorphic residues. The hot spots in the proteins under study were identified by KFC2 [36]. The analyses are based on the structural microenvironment around amino acid residues and take into consideration hot spots determined experimentally. The hot spot prediction takes into account conformation specificity (K-FADE score) and biochemical features such as hydrophobicity, polar and electrostatic interactions (K-CON score) through a machine learning approach. This approach identifies amino acid residues that significantly contribute to the binding free energy within the protein-protein interaction interface. Clinically important polymorphic residues for the TRAF5 protein were identified through the dbSNP (database of single nucleotide polymorphism; https://www.ncbi.nlm.nih. gov/SNP).
Results and Discussion
H. sapiens TRAF5 structure organization
TRAF5 is a protein that takes parts in several biological process, including activation of immune system cells [37]. At least three main factors guarantee the functions performed by the protein: the stability of its three-dimensional conformation, the presence of conserved domains and the protein partners that TRAF5 interacts with (Figure 1). TRAF5 contains three zinc-finger motifs, which form finger-like projections in order to make interact with target molecules [38]. A zinc-finger motif often has plastic forms of binding, with active roles on gene transcription and translation [39], mRNA transport [40], cytoskeleton arrangement [41], epithelial development [42], cell adhesion [43], protein folding and interaction with partner [44] and chromatin remodeling [45]. The zinc-finger domain can also regulate several aspects of the immune system roles against pathogens. Certain proteins with this motif act targeting mRNAs and proteins for degradation, consequently controlling signaling pathways in immune response via cytokine synthesis and activation of immune cells [46]. Thus, proteins containing this domain play roles in regulating ubiquitination processes [47,48]. The three zinc-fingers found within TRAF5 structure are located at amino acid residues 45- 85, 127-181 and 182-239.
TRAF5 also contain a MATH or TRAF domain in the C-terminus of the protein [49], from the 403 amino acid residue to the 549. This domain participates in selfassembly and interaction performed with other protein partners [50]. The domain forms an antiparallel beta sandwich structure. TRAF5 presents a coiled-coil motif (residues 237-342) near to the MATH domain; this conformation permits proper interaction with targets to form complexes that signals for the TNF receptor-1 during immune response events [51]. Thus, TRAF5 mediates activation of NF-kappa-B (nuclear factor kappa B) and JNK (Jun N-terminal kinases) through these domains.
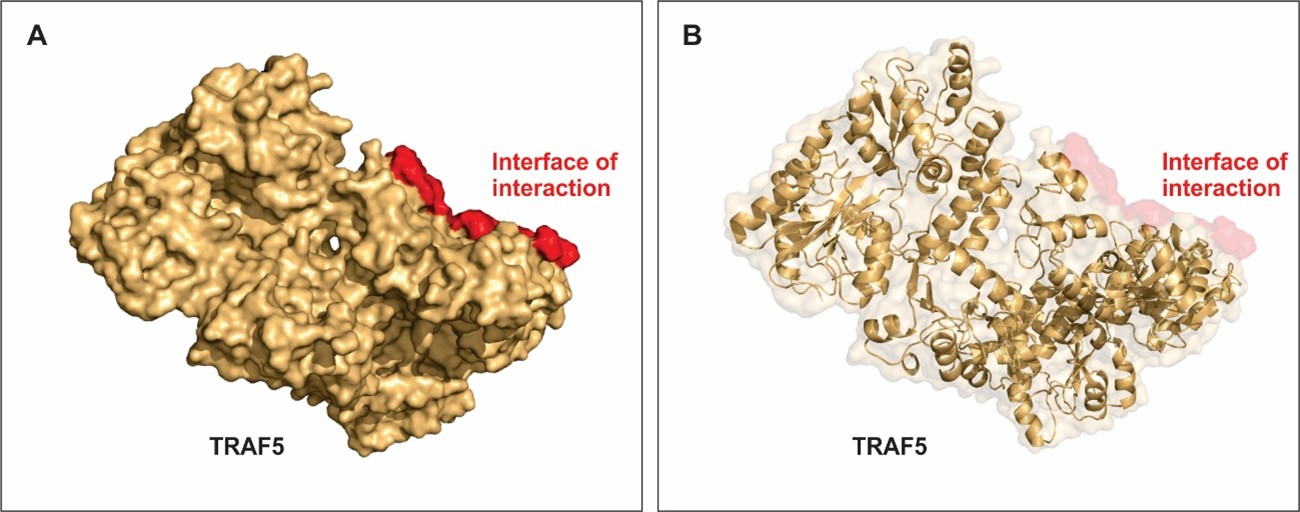
Approximately 22% of the TRAF5 structure has helical shape and around 31% is beta-sheet (Figure 2). Figure 2A shows the surface structure of TRAF 5 and figure 2B shows details on the secondary structure with helices, alfa sheets and loops. Figure 2A and B also shows the region where interaction with E6 from HPV is most stable. Important residues within this region accounts for the low free energy of binding between these host and pathogen proteins. The docking results showed that best cluster, consequently with the lowest energy, had a score of -791.7 (data not shown). The results and scientific evidences support our model predicted for the conformational structure of TRAF5 complexes with HPV E6.
HPV E6 structure organization
HPV infection induces host cells immortalization, which contributes immune response evasion and cancer development. Two main HPV genes stay hidden within host genome, making cells expresses the oncogenic proteins they code for. Over expression of E6 and E7 maintains the host cells immortalization, regulates cell phenotype and malignancy transformation and the consequent loss of mitosis control [52]. E6 and E7 are highly expressed in tumor cervical cells, alter the synthesis and activity of cytokines and host tumor suppressor proteins, such as p53 [53].
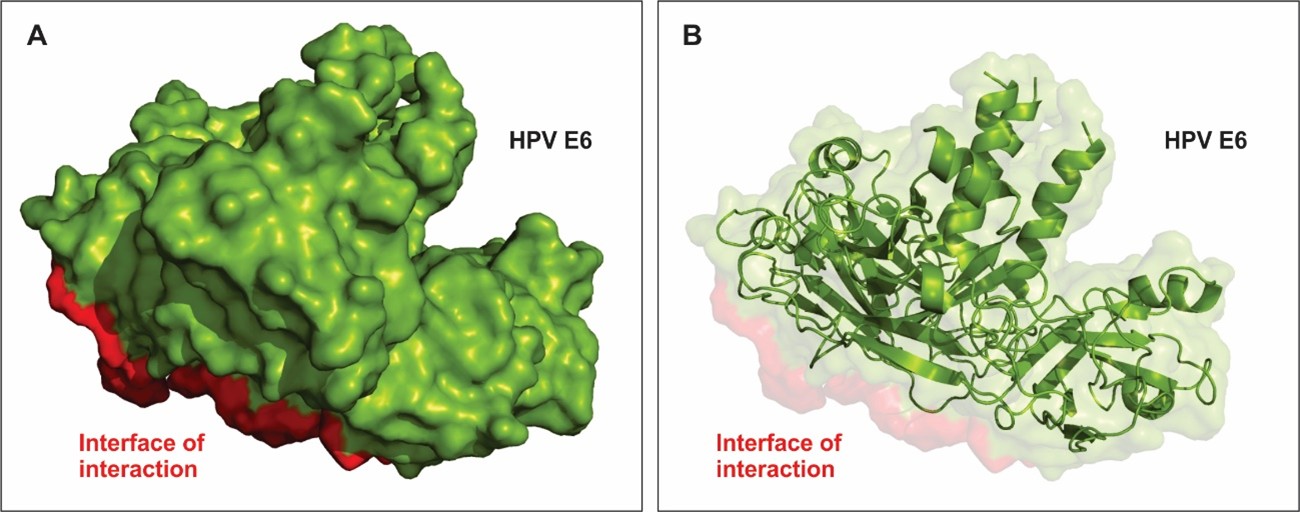
E6 protein contains domain named E6 superfamily and a conserved zinc-binding motif. Through this motif, E6 is able to interfere with the metabolism host proteins such as those related to immune response (TRAF family) [29] and those that control cell cycle, such as p53 [53]. Approximately 46% of E6 structural are helical spanning through 176 amino acid residues and 20% beta-sheet spanning through 79 residues. Figure 3A shows the most stable conformational surface structure of HPV E6 when bound to TRAF5 and their interface of interaction. Figure 3B shows the pattern of alfa-helices and beta-strands that comprise the secondary structure of the protein.
Interaction between TRAF5 and HPV E6
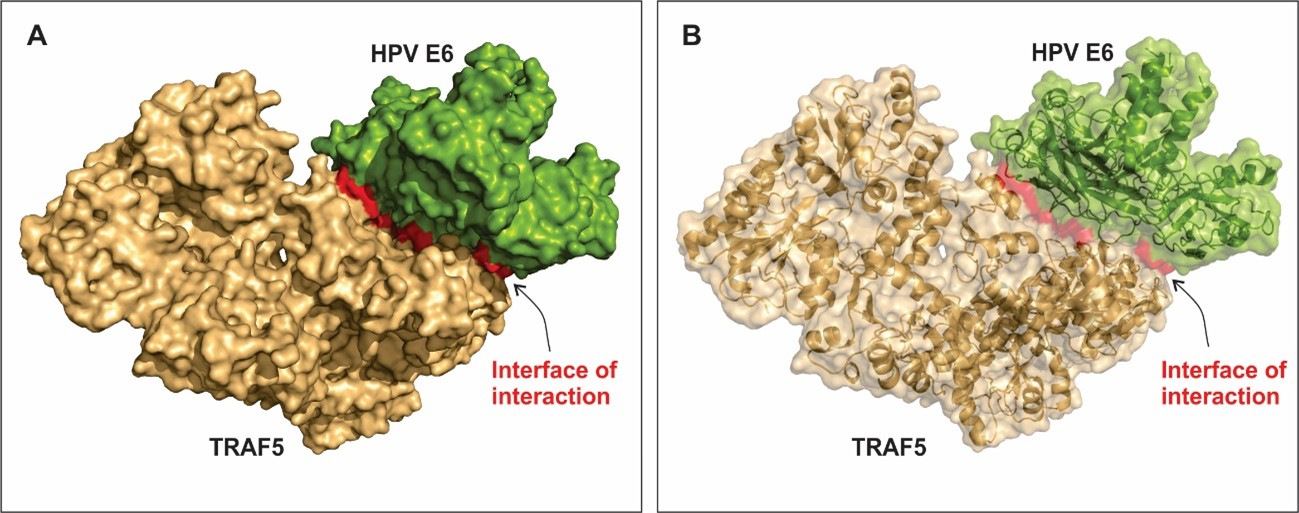
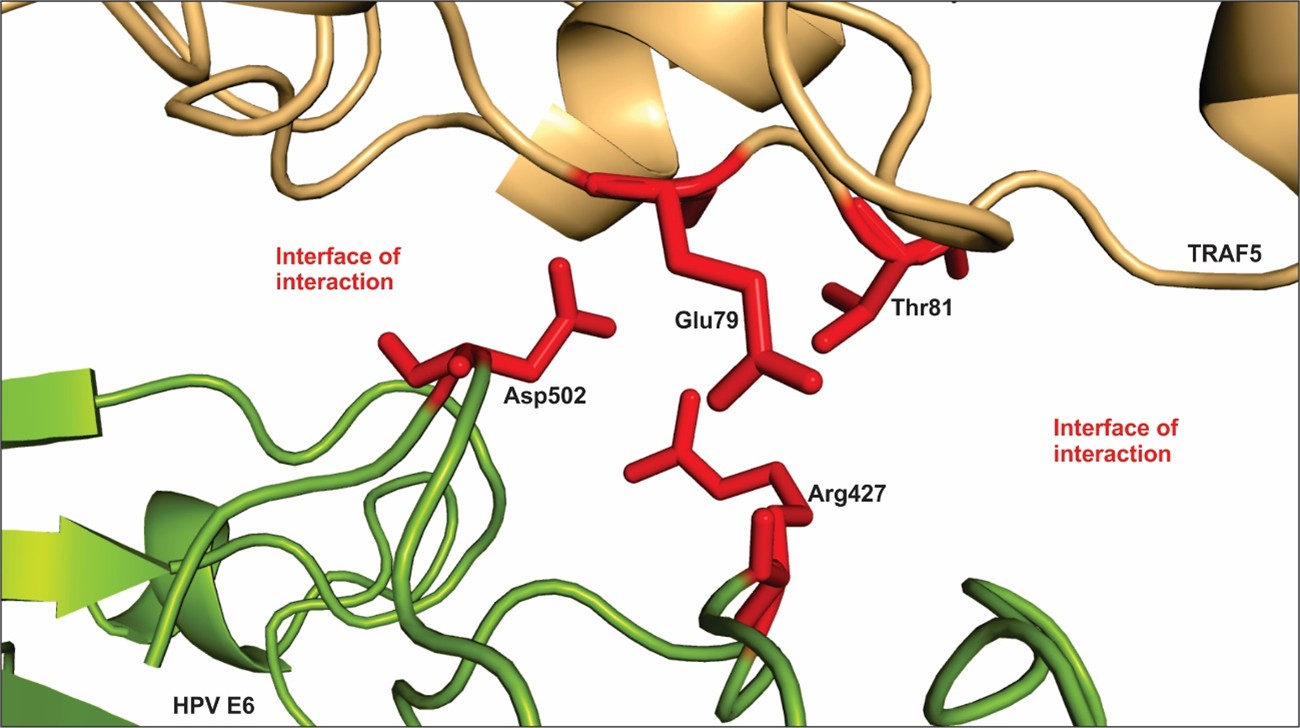
Figure 4A and B shows the best model of interaction between TRAF5 and HPV E6. Amino acid residues scored as hot spots stabilize the large interface of interaction between the bindings of host-pathogen proteins. The residues that most contribute to free-energy of binding are listed on table 1. The hot spot residues are place within the interface of interaction (Figure 4) and they interact with amino acid from the other protein through polar, hydrophobic and electrostatic interactions. Interestingly, all the residues that most contribute to the stabilization of the complex are polar amino acids.
| Protein | Chain | Residue | HP score* |
|---|---|---|---|
| E6 | A | Glu79 | 0.15 |
| E6 | A | Thr81 | 0.13 |
| E6 | C | Gln14 | 0.28 |
| TRAF5 | b | Arg427 | 0.04 |
| TRAF5 | c | Arg427 | 0.08 |
| TRAF5 | c | Asp502 | 0.86 |
Polar interactions are key contributors to the specificity of interactions. Here, we hypothesize that for high-risk HPV types (such as HPV 16), the complex formed by E6 and TRAF5 are well stabilized mainly by those residues (Table 1) and this interaction my reduce TRAF5 activity. Therefore, it alters the levels of cytokines for a proper immune response.
Figure 5 shows a small segment of the interface of interaction between the proteins TRAF5 and E6, with the hotspot residues listed on table 1. The amino acid residues Thr81 and Glu79 located on the TRAF5 sequence and the residues Arg427 and Asp502 located on the E6 sequence project into the interface of interaction and contribute to the free-energy of the interaction. Neighbor residues also contribute to stabilize the complex. E6 and other oncogenes from high-risk HPV types deregulate immune response and might lead to malignancy transformation of the host cells [52].
The results from our in silico approaches showed the most stable model for the interaction between E6 and TRAF5, then we chose a region of the interface of interaction which contains several hot spot residues (Table 1) in order to rationally design a inhibiting peptide against this interaction. Other approaches have designed peptides against HPV [54,55] for the development of vaccines and alternative treatment trials. We designed a peptide containing 21 amino acid residu that interacts with the E6 protein in order to modulate its function and inactivate it (Figure 6).
The rational construction of peptides based on structure explores feasible strategies for new therapies that induce proper immune response in order to combat HPV infections efficiently.
Concluding remarks
HPV is the most common sexual transmitted disease worldwide. It is a complex and expensive public health problem. The disease has a high rate of deaths due to its relation to cancer, principally cervix cancer. Several HPV genes and proteins are potential candidates as genetic markers for the development of cancer and they are target for vaccines and HPV treatment.
Here, we hypothesize that interaction between TRAF5 and E6 could be modulated in order to inhibit the activity of E6. We have shown an in silico approach of interaction between TRAF5 and HPV E6 through the identification of hot spots within the interface of interaction of the complex. We propose a new peptide that interacts and inhibits HPV E6. For a future perspective, the peptide designed will be tested in vitro and other regions of the interface of interaction will also be screened so that other peptides could also be designed and tested.
References
2. Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Bro- ker TR, Stanley MA. The biology and life-cycle of human papillomaviruses. Vaccine. 2012 Nov 20;30:F55-70.
3. Insinga RP, Perez G, Wheeler CM, Koutsky LA, Gar- land SM, Leodolter S, Joura EA, Ferris DG, Steben M, Hernandez-Avila M, Brown DR. Incident cervical HPV infections in young women: transition probabilities for CIN and infection clearance. Cancer Epidemiology and Prevention Biomarkers. 2011 Feb 1;20(2):287-96.
4. STD Facts (2012) - Human papillomavirus (HPV).
5. Gilbert LK, Alexander L, Grosshans JF, Jolley L. An- swering frequently asked questions about HPV. Sexually transmitted diseases. 2003 Mar 1;30(3):193-4.
6. Ho GY, Bierman R, Beardsley L, Chang CJ, Burk RD. Natural history of cervicovaginal papillomavirus infection in young women. New England Journal of Medicine. 1998 Feb 12;338(7):423-8.
7. Ljubojevic S, Skerlev M. HPV-associated diseases. Clinics in dermatology. 2014 Mar 1;32(2):227-34.
8. Schiller JT, Day PM, Kines RC. Current understanding of the mechanism of HPV infection. Gynecologic oncology. 2010 Jun 1;118(1):S12-7.
9. Woodham AW, Da Silva DM, Skeate JG, Raff AB, Ambroso MR, Brand HE, Isas JM, Langen R, Kast WM. The S100A10 subunit of the annexin A2 heterotetramer facilitates L2-mediated human papillomavirus infection. PloS one. 2012 Aug 22;7(8):e43519.
10. Pahud BA, Ault KA. The expanded impact of human papillomavirus vaccine. Infectious Disease Clinics. 2015 Dec 1;29(4):715-24.
11. SCHEURER ME, Tortolero-Luna G, Adler-Storthz K. Human papillomavirus infection: biology, epidemiology, and prevention. International Journal of Gynecologic Cancer. 2005 Sep 8;15(5):727-46.
12. Tan S, de Vries EGE, van der Zee AGJ, de Jong S. Anticancer drugs aimed at E6 and E7 activity in HPV-positive cervical cancer. Curr Cancer Drug Targets. 12(2), 170–184 (2012).
13. Garzetti G, Ciavattini A, Goteri G, de Nictolis M, Menso S, Muzzioli M, Fabris N. HPV DNA positivity and natural killer cell activity in the clinical outcome of mild cervical dysplasia: integration between virus and immune system. Gynecologic and obstetric investigation. 1995;39(2):130-5.
14. Malejczyk J, Malejczyk M, Köck A, Urbanski A, Majewski S, Hunzelmann N, Jablonska S, Orth G, Luger TA. Autocrine growth limitation of human papillomavirus type 16-harboring keratinocytes by constitutively released tumor necrosis factor-alpha. The Journal of Immunology. 1992 Oct 15;149(8):2702-8.
15. Woodworth CD, McMullin E, Iglesias M, Plowman GD. Interleukin 1 alpha and tumor necrosis factor alpha stimulate autocrine amphiregulin expression and proliferation of human papillomavirus-immortalized and carcinoma-derived cervical epithelial cells. Proceedings of the National Academy of Sciences. 1995 Mar 28;92(7):2840-4.
16. Kyo S, Inoue M, Hayasaka N, Inoue T, Yutsudo M, Tanizawa O, Hakura A. Regulation of early gene expression of human papillomavirus type 16 by inflammatory cytokines. Virology. 1994 Apr 1;200(1):130-9.
17. Chagas BS, Gurgel AP, Paiva Júnior SS, Lima RC, Cordeiro MN, Moura RR. Synergic effect of oral contraceptives, GSTP1 polymorphisms, and high-risk HPV infection in development of cervical lesions. Genet Mol Res. 2017 Aug 17;16(3).
18. McCusker C, Upton J, Warrington R. Primary immunodeficiency. Allergy, Asthma & Clinical Immunology. 2018 Sep;14(2):61.
19. Boscolo-Rizzo P, Furlan C, Lupato V, Polesel J, Fratta E. Novel insights into epigenetic drivers of oropharyngeal squamous cell carcinoma: role of HPV and lifestyle factors. Clinical epigenetics. 2017 Dec;9(1):124.
20. Sudenga SL, Shrestha S, Macaluso M, Partridge EE, Johanning GL, Piyathilake CJ. Functional variants in CYP1A1 and GSTM1 are associated with clearance of cervical HPV infection. Gynecologic oncology. 2014 Dec 1;135(3):560-4.
21. Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nature Reviews Immunology. 2009 Oct;9(10):679.
22. Cornet I, Bouvard V, Campo MS, Thomas M, Banks L, Gissmann L, Lamartine J, Sylla BS, Accardi R, Tommasino M. Comparative analysis of transforming properties of E6 and E7 from different beta human papillomavirus types. Journal of virology. 2012 Feb 15;86(4):2366-70.
23. Spangle JM, Münger K. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. Journal of virology. 2010 Sep 15;84(18):9398-407.
24. Brenna SM, Syrjänen KJ. Regulation of cell cycles is of key importance in human papillomavirus (HPV)-associated cervical carcinogenesis. São Paulo Medical Journal. 2003;121(3):128-32.
25. Liu X, Roberts J, Dakic A, Zhang Y, Schlegel R. HPV E7 contributes to the telomerase activity of immortalized and tumorigenic cells and augments E6-induced hTERT promoter function. Virology. 2008 Jun 5;375(2):611-23.
26. Prati B, Marangoni B, Boccardo E. Human papillomavirus and genome instability: from productive infection to cancer. Clinics. 2018;73.
27. Ronchi VP, Klein JM, Edwards DJ, Haas AL. The active form of E6-associated protein (E6AP)/UBE3A ubiquitin ligase is an oligomer. Journal of Biological Chemistry. 2014 Jan 10;289(2):1033-48.
28. Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztányi Z, El-Gebali S, Fraser M, Gough J. InterPro in 2017—beyond protein family and domain annotations. Nucleic acids research. 2016 Nov 28;45(D1):D190-9.
29. Boulabiar M, Boubaker S, Favre M, Demeret C. Keratinocyte sensitization to tumour necrosis factor-induced nuclear factor kappa B activation by the E2 regulatory protein of human papillomaviruses. Journal of General Virology. 2011 Oct 1;92(10):2422-7.
30. Vuillier F, Gaud G, Guillemot D, Commere PH, Pons C, Favre M. Loss of the HPV-infection resistance EVER2 protein impairs NF-κB signaling pathways in keratinocytes. PLoS One. 2014 Feb 19;9(2):e89479.
31. Ghoorah AW, Devignes MD, Smaïl-Tabbone M, Ritchie DW. KBDOCK 2013: a spatial classification of 3D protein domain family interactions. Nucleic acids research. 2013 Nov 23;42(D1):D389-95.
32. Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, Gough J. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic acids research. 2018 Nov 6;47(D1):D351-60.
33. Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O’Donnell L, Oster S, Theesfeld C, Sellam A, Stark C. The BioGRID interaction database: 2017 update. Nucleic acids research. 2017 Jan 4;45(D1):D369-79.
34. Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S. The ClusPro web server for protein–protein docking. Nature protocols. 2017 Feb;12(2):255.
35. Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins: Structure, Function, and Bioinformatics. 2006 Nov 1;65(2):392-406.
36. Darnell SJ, Page D, Mitchell JC. An automated decision- tree approach to predicting protein interaction hot spots. Proteins: Structure, Function, and Bioinformatics. 2007 Sep;68(4):813-23.
37. Shen J, Qiao YQ, Ran ZH, Wang TR. Up-regulation and pre-activation of TRAF3 and TRAF5 in inflammatory bowel disease. International journal of medical sciences. 2013;10(2):156.
38. Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 32(2), 63–70 (2007).
39. Klug A. Zinc finger peptides for the regulation of gene expression. Journal of molecular biology. 1999 Oct 22;293(2):215-8.
40. Pohlmann T, Baumann S, Haag C, Albrecht M, Feldbrügge M. A FYVE zinc finger domain protein specifically links mRNA transport to endosome trafficking. Elife. 2015 May 18;4:e06041.
41. Turkel N, Portela M, Poon C, Li J, Brumby AM, Richardson HE. Cooperation of the BTB-Zinc finger protein, Abrupt, with cytoskeletal regulators in Drosophila epithelial tumorigenesis. Biology open. 2015 Aug 15;4(8):1024-39.
42. Singh J, Itahana Y, Parrinello S, Murata K, Desprez PY. Molecular cloning and characterization of a zinc finger protein involved in Id-1-stimulated mammary epithelial cell growth. Journal of Biological Chemistry. 2001 Apr 13;276(15):11852-8.
43. Beckerle MC. Zyxin: zinc fingers at sites of cell adhesion. Bioessays. 1997 Nov;19(11):949-57.
44. Leon O, Roth M. Zinc fingers: DNA binding and protein-protein interactions. Biological research. 2000;33(1):21-30.
45. Ravasi T, Huber T, Zavolan M, Forrest A, Gaasterland T, Grimmond S, Hume DA, Riken GER Group. Systematic characterization of the zinc-finger-containing proteins in the mouse transcriptome. Genome research. 2003 Jun 1;13(6b):1430-42.
46. Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, Akira S. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009 Apr;458(7242):1185.
47. Fu M, Blackshear PJ. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nature Reviews Immunology. 2017 Feb;17(2):130.
48. Wang X, Yang J, Han L, Zhao K, Wu Q, Bao L, Li Z, Lv L, Li B. TRAF5-mediated Lys-63-linked polyubiquitination plays an essential role in positive regulation of RORγt in promoting IL-17A expression. Journal of Biological Chemistry. 2015 Nov 27;290(48):29086-94.
49. Sunnerhagen M, Pursglove S, Fladvad M. The new MATH: homology suggests shared binding surfaces in meprin tetramers and TRAF trimers. FEBS letters. 2002 Oct 23;530(1-3):1-3.
50. Ye H, Park YC, Kreishman M, Kieff E, Wu H. The structural basis for the recognition of diverse receptor sequences by TRAF2. Molecular cell. 1999 Sep 1;4(3):321- 30.
51. Häcker H, Tseng PH, Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nature Reviews Immunology. 2011 Jul;11(7):457.
52. Androphy EJ, Hubbert NL, Schiller JT, Lowy DR. Identification of the HPV-16 E6 protein from transformed mouse cells and human cervical carcinoma cell lines. The EMBO journal. 1987 Apr 1;6(4):989-92.
53. Thomas MC, Chiang CM. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Molecular cell. 2005 Jan 21;17(2):251-64.
54. Nilges K, Höhn H, Pilch H, Neukirch C, Freitag K, Talbot PJ, Maeurer MJ. Human papillomavirus type 16 E7 peptide-directed CD8+ T cells from patients with cervical cancer are cross-reactive with the coronavirus NS2 protein. Journal of virology. 2003 May 1;77(9):5464-74.
55. Liu TY, M Hussein W, Toth I, Skwarczynski M. Advances in peptide-based human papillomavirus therapeutic vaccines. Current topics in medicinal chemistry. 2012 Jul 1;12(14):1581-92.