Commentary
Engagement of mature T cell receptor (TCR), a multiprotein complex consisting of an αβ heterodimer associated with invariant CD3 signaling proteins, on CD4+CD8+ double positive (DP) thymocytes by selfpeptide/ self-MHC complex on thymic stromal cells results in negative selection of thymocytes expressing strong affinity TCRs and positive selection of thymocytes expressing weak affinity TCRs. Positively selected thymocytes almost invariably differentiate into MHCIIspecific CD4+ helper and MHCI-specific CD8+ cytotoxic T cells endowed with effector functions crucial for effective cell-mediated immune responses [1,2]. Together, thymic selection and lineage choice ensures generation of mature T cell repertoire that is capable of mounting effective responses to non-self-antigens but is tolerant to selfantigens encountered in the periphery [3,4]. Although correlation between MHC specificity with helper and cytotoxic lineage choice remains unclear, it appears to be influenced by duration and intensity of TCR signaling that imprints transcriptional program underlying the two lineages [5]. Specificity of CD4 and CD8 coreceptor for MHCII and MHCI, respectively, is important in TCR signaling with CD4, compared to CD8, promoting stronger signaling due to stronger association of Src tyrosine kinase Lck with cytoplasmic tail of CD4 than that of CD8 [6-10]. Irrespective of MHC specificity positively selected DP thymocytes temporally downregulate Cd8 transcription and become CD4+CD8lo thymocytes that eventually develop into CD4+ and CD8+ single positive (SP) mature thymocytes [3,11]. Indeed, ablating CD4 expression at CD4+CD8lo stage redirects positively selected thymocytes into the cytotoxic lineage [12]. Thus, kinetics of coreceptor, particularly CD4, expression plays an important role in CD4 vs CD8 lineage choice. Based on these observations kinetic signal strength model has been proposed for CD4/CD8 lineage choice [13]. Accordingly, in CD4+CD8lo thymocytes downregulation of CD8 results in reduced Lck activity culminating in shorter or weaker signal transduction in MHCI-signaled thymocytes, while continued CD4 expression at this stage, and therefore higher Lck activity, results in sustained or stronger signal in MHCII-signaled thymocytes. However, two observations suggest that kinetic signal strength model alone cannot explain the CD4 helper versus CD8 cytotoxic lineage choice; (a) constitutive CD8 or chimeric CD8.4 (cytoplasmic domain of CD8 substituted by that of CD4) redirects only a fraction of MHCI-signaled thymocytes into CD4 lineage [14] and (b) positively selected MHCI-restricted thymocytes expressing strong affinity TCR such as OTI- TCR and P14- TCR, which transduce stronger signal compared to several MHCII-specific TCRs such as OTII-TCR, fail to develop into the CD4 helper lineage [15]. It is proposed that signal disruption renders MHCI-signaled thymocytes sensitive to cytokine signaling critical for Runx3 activation and the CD8 cytotoxic lineage choice [16,17], while stronger TCR signal induces ThPOK (Zbtb7b, cKrox) in MHCII- signal thymocytes essential for the CD4 helper lineage choice [18]. Identification of Helper Deficient (HD) mouse and subsequent research showed that loss and gain of ThPOK function redirects, respectively, MHCII-specific cells into the CD8 cytotoxic lineage and MHCI-specific cells into the CD4 helper lineage [19-22]. In contrast, Runx3 deficiency or constitutive expression alters lineage choice of only a fraction of MHCI-signaled thymocytes [23-26]. Collectively, these observations suggest a dominant role for ThPOK in the CD4 helper versus CD8 cytotoxic lineage choice of developing thymocytes.
Although MHC specificity of positively selected thymocytes correlates with functionally distinct lineages, cellular components associated with proximal TCR signaling are similar except for stronger Lck association with the cytoplasmic tail of CD4 than that of CD8. However, higher Lck activity in MHCII-signaled cells alone cannot explain CD4 lineage choice as constitutively active or dominant negative Lck alters lineage fate of only a small number of positively selected thymocytes expressing monoclonal TCR [8,9]. Similarly, temporal and differential kinetics of ZAP70 expression, delayed and higher expression in the CD8 compared to CD4 committed thymocytes, is proposed to play a role in lineage divergence although mechanism of its action remains undefined [5,27-29]. How differences in kinetics and/or strength of proximal TCR signaling lead to activation of lineage specifying ThPOK and Runx3 expression in MHCII- and MHCI-signaled thymocytes, respectively, is challenging and remains to be elucidated.
Central role for ThPOK in defining the CD4 versus CD8 lineage divergence has prompted intense investigation of this BTB/POZ zinc finger transcription factor over the last decade. Previous studies showed that constitutive ThPOK expression redirects MHCI-signaled thymocytes into the CD4 helper lineage [20,30]; however it remained unclear if a defined amount of ThPOK promoted the CD4 lineage choice of MHCI- and MHCII-signaled thymocytes at comparable efficiency and whether TCR signal strength or MHC specificity played any role in this process. In our recently published article, we evaluated role of ThPOK dose and TCR signal strength on the CD4 lineage choice of MHCI- and MHCII-signaled thymocytes [31]. Analysis of three independent transgenic mouse lines expressing different amounts of ThPOK in developing thymocytes showed that the efficiency of CD4 lineage choice of MHCIsignaled thymocytes expressing monoclonal (OTI- and P14-TCR) or polyclonal (MHCII-/- mice) TCR specificities was directly proportional to ThPOK levels in the preselection DP thymocytes. These data suggest the extent of transgenic ThPOK occupancy at the target gene loci may prime positively selected MHCI-signaled thymocytes for the CD4 lineage development. The ability of retrogenic ThPOK expression to only partially reprogram peripheral CD8 cytotoxic T cells into the CD4 helper T cell supports this possibility [30]. Interestingly, all three transgenic lines expressed higher amount of ThPOK compared to endogenous ThPOK levels observed in CD4+ T cells from WT mice, and yet the lowest ThPOK expressing transgene (ThPOK-H transgene) caused partial CD8 to CD4 lineage redirection. As expected Runx3 was suppressed in the redirected MHCI-specific CD4+ T cells. Interestingly, a substantial number of MHCI-specific mature T cells in one of the ThPOK transgenic line failed to activate/sustain CD4 expression leading to production of mature DN T cells in these mice. This was despite suppression of Runx3 which silences Cd4 in CD8+ T cells [26,32] indicating complex regulation of Cd4 [33,34]. Lineage associated gene expression analysis showed suppression of cytotoxic program and failure of induction of helper program in DN T cells in ThPOK transgenic mice. The absence of mature DN T cells in the thymus and in vitro differentiation of CD4+ SP thymocytes into DN T cells suggest a complex regulation of Cd4 expression requiring sustained TCR signaling in developing thymocytes [33,34]. Our subsequent analysis showed that the same ThPOK transgene completely restores CD4 development of MHCII-specific OTII-TCR in Thpok-/- mice providing first indication that, compared to MHCII-signaled thymocytes, MHCI-signaled thymocytes require significantly higher amount of ThPOK for efficient CD4 lineage choice. Further experimentations showed that augmenting TCR signal strength via introduction of constitutively active Lck increases the efficiency of ThPOKinduced CD4 lineage choice of MHCI-specific thymocytes although it was still lower compared to the CD4 lineage choice of MHCII-signaled thymocytes expressing the same amount of ThPOK. Together these analyses suggest that suppression of cytotoxic and induction of helper program require different amount of ThPOK, and strength and quality of TCR signaling largely determines the efficiency of ThPOK-induced CD4 lineage choice.
How might TCR signal strength and MHC specificity segregate CD4 versus CD8 lineage choice? Mechanistically there are two mutually nonexclusive possibilities that may underlie differential impact of a defined amount of ThPOK on lineage choice. It is conceivable that during a temporal window of lineage choice [35], accessibility of genes regulated by ThPOK or Runx3 is influenced by strength and duration of TCR signaling; stronger and longer duration of TCR signal in MHCII-signaled thymocytes may permit target gene loci access for an extended period requiring a smaller amount of ThPOK for modulating their expression leading to suppression of cytotoxic program and induction of helper program. Conversely, TCR signal disruption in MHCI-signaled thymocytes reduced accessibility of target genes would require significantly more ThPOK (than the amount required for CD4 lineage choice of MHCII-signaled thymocytes) for the CD4 helper lineage choice. Increased efficiency of CD4 lineage choice of MHCI-specific thymocytes due to augmented TCR signaling in ThPOK transgenic mice supports this notion [31]. Such a temporal target gene accessibility model may explain incomplete modulation of helper and cytotoxic lineage genes in peripheral CD8 cytotoxic T cells following enforced ThPOK expression [30] or in developing thymocytes with impaired Runx function due to compound deficiency of Tel/Groucho proteins (Tle1/3/4) or MAZR and Runx3 leading to the production of mature T cells expressing both CD4 and CD8 coreceptors with undefined functional potential [36-38]. We propose that strong/sustained TCR signaling during a temporal window alters the chromatin structure allowing longer accessibility of genes essential for the induction of CD4 helper lineage program and suppression of CD8 cytotoxic lineage program. Such a model also explains the continued presence of regulatory T cells, albeit reduced in number, in ThPOK-deficient mice as generation of these cells requires high affinity TCR interaction with agonist selfpeptide/ MHC in the thymus [39]. An alternate attractive possibility could be competition between ThPOK and Runx3 for the same set of genes involved in lineage choice, which may explain the generation of CD4+T cells in mice lacking ThPOK and Runx protein function [39]. However, kinetic signal strength model alone cannot explain why the same ThPOK transgene promotes the CD4 lineage choice of MHCII-specific thymocytes more efficiently than that of MHCI-specific thymocytes with augmented TCR signaling. In fact, the strength of TCR signaling in MHCIrestricted thymocytes expressing constitutive active Lck was greater than MHCII-specific thymocytes and yet, the efficiency of CD4 lineage choice due to defined amount of ThPOK was significantly different [31]. This suggests a role for quantitatively as well as qualitatively TCR signaling in CD4 helper versus CD8 cytotoxic lineage choice. While higher Lck activity mimics quantitative aspect of TCR signaling, how qualitative signaling (i.e., MHC specificity) influences CD4 versus CD8 lineage choice remains unclear. It is plausible that in MHCII-signaled thymocytes the qualitative TCR signaling may open up the Thpok and its target gene loci for modifications by chromatin modifiers [33,40-43], while quantitative TCR signaling may enhance the temporal accessibility allowing sufficiently high expression of these genes and CD4 lineage choice.
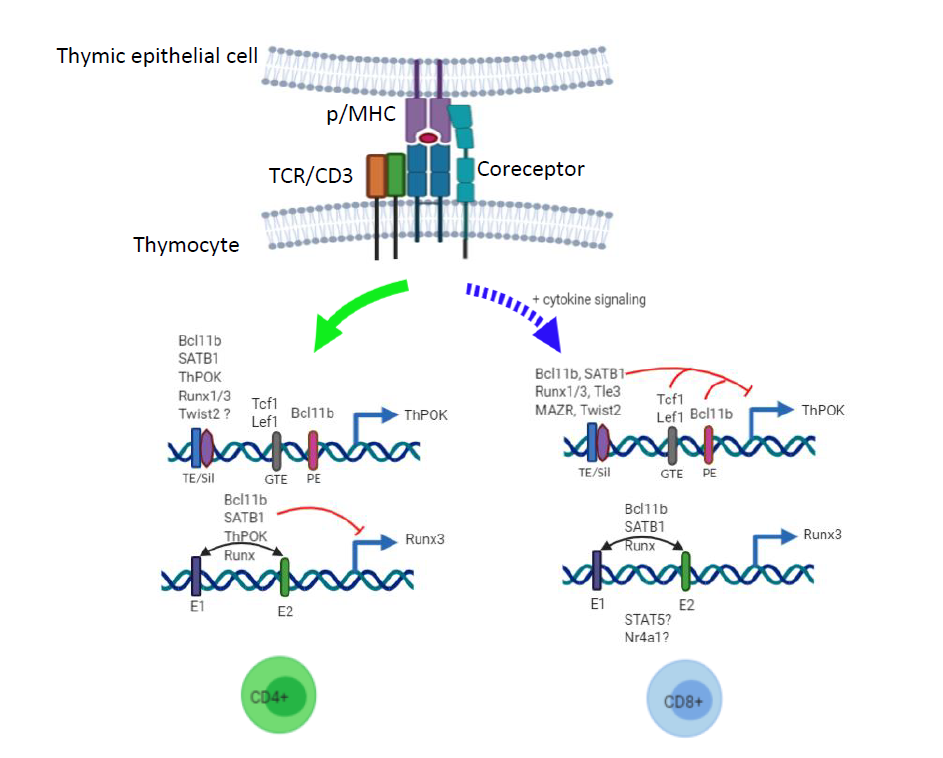
ThPOK is a master regulator of CD4 helper versus CD8 cytotoxic lineage choice, and therefore it is pertinent to understand how it is induced in MHCII-signaled thymocytes and repressed in the preselection DP and MHCI-signaled thymocytes. Transcription of Thpok from the distal P1 and the proximal P2 promoters is regulated by two major cis acting motifs, a distal regulatory element (DRE) located about 15kb upstream and a proximal enhancer (PE) located at about 4.5kb upstream of the coding exon [38,44]. A third regulatory motif called general T cell element (GTE) of unknown function lies between the silencer and PE motifs. The DRE consists of a thymic enhancer (TE) and a silencer motif (Sth). TE regulates ThPOK expression in positively selected thymocytes and PE further augments, mainly via P2 activity, ThPOK levels during the subsequent stages of CD4 lineage choice [19,35,45]. Bcl11b, a zinc finger transcription factor, represses ThPOK expression in the silencer-independent and - dependent manner, respectively, in preselection thymocytes and CD8+ T cells, while it promotes ThPOK expression in CD4+ T cells by augmenting enhancer activity [42]. SATB1, a chromatin organizer, regulates Thpok transcription by modulating TE, PE and Sth activities [41]. Interestingly, both these factors are reported to influence Runx3 expression as well. It is suggested that Bcl11b and SATB1 prime lineage specifying genes in the preselection thymocytes and upon TCR signaling allow recruitment of other factors critical for ThPOK or Runx3 expression leading to commitment of the selected thymocytes into relevant functional lineages (Figure 1). Specifically, it may result in induction of GATA3 and TOX expression in MHCII-signaled thymocytes both of which are reported to act upstream of ThPOK and are critical for the CD4 lineage choice [46,47]. ThPOK expression, with some contribution from GATA3, suppresses the cytotoxic lineage program primarily by suppressing Runx3 expression [48]. Other factors known to regulate Thpok induction are the teneleven translocation (TET) DNA demethylation proteins probably via DNA demethylation of TE and PE motifs [49,50]. How ThPOK commits signaled thymocytes and maintains the CD4 helper lineage remains an area of active research. ThPOK positively regulates members of Socs family; in Thpok-deficient mice transgenic SOCS1 substantially restores CD4 lineage choice and in ThPOK transgenic mice SOCS1 deficiency partially restores the CD8 cytotoxic lineage development due to Runx3 expression [51]. Interestingly, the rescue of CD4 lineage development in SOCS1 transgenic mice required MHCIIsignaling supporting a role for qualitative TCR signaling in CD4 versus CD8 lineage choice. SOCS1 deficiency alone redirects a small number of MHCII-signaled thymocytes into the CD8 cytotoxic lineage, and it would be interesting to determine if these cells continue to express ThPOK [51,52].
In MHCI-signaled thymocytes Runx3 induction is critical in suppressing ThPOK expression and promoting cytotoxic lineage choice. Runx proteins suppress Thpok induction by regulating the silencer activity in the preselection and CD8 committed thymocytes [38]. Interestingly, Runx protein complexes are associated with the Thpok silencer and yet fail to suppress ThPOK expression in CD4+ T cells [38]. This suggests that Runx proteins likely recruit other factors that together repress Thpok induction in preselection and CD8 SP thymocytes. Indeed, Tle/ Groucho transcriptional corepressors, particularly Tle3, inhibit Thpok induction in MHCI-signaled thymocytes in Runx protein dependent manner [36]. Interestingly, Tle proteins also prevents expression of the helper lineage genes such as Cd4 and St8sia6 via its association with HMG group transcription factor Tcf1. Tcf1 and Lef1, another HMG group transcription factor, also bind to GTE and suppress Thpok induction via intrinsic histone deacetylase activity in the CD8 committed cells [40]. A recent report show that enforced Twist2 expression significantly impairs the CD4 T cell development and redirects some of them into the CD8 lineage. Interestingly, Twist2 deficiency moderately impairs the CD8+ mature T cell production but fails to redirect them into the CD4 lineage [53]. Transgenic Twist2 significantly suppressed Thpok induction in CD4+CD8lo thymocytes and Twist2 deficiency moderately upregulated ThPOK levels in CD8 SP thymocytes although it does not appear to be sufficient for Runx3 suppression and promotion of the CD4 lineage choice of MHCI-signaled thymocytes. While inhibition of ThPOK expression MHCI-signaled thymocytes is known in great detail, the mechanism of Runx3 induction in these cells remains unclear. In the CD8 committed thymocytes Runx3 is expressed mainly from the distal promoter P1 via at least two enhancer motifs located 21kb and 39kb upstream of the P1 promoter [41], and ChIP-seq analysis showed association of Runx, SATB1, Bcl11b and ThPOK with these enhancers [41,42] (Figure 1). However, the mechanism of activation of Runx3 enhancers in the CD8 committed thymocytes remains unclear. As cytokine signaling plays an important role in Runx3 expression and CD8 lineage choice, it is conceivable that Stat5, a mediator of γc cytokine signaling, may bind to these enhancers and activate Runx3 induction [17]. SATB1 or Bcl11b are proposed to induce Runx3 and suppress Thpok in the CD8 committed thymocytes; however, as mentioned above SATB1 and Bcl11b suppress Runx3 and induce Thpok in the CD4 committed thymocytes. Thus, SATB1 and Bcl11b likely alter chromatin structure allowing recruitment of putative factors leading to induction of lineage specific ThPOK and Runx3 expression that matches with MHC specificity of the signaled thymocytes. Finally, Nur77 (Nr4a1), in association with CoREST corepressor, negatively regulates Runx3 expression [54] although it is not clear if this complex binds to Runx3 enhancers. Interestingly, Nur77 deficiency impairs the CD8 T cell development without promoting the CD4 helper lineage choice [54]. This observation also supports the idea that TCR and p/MHC interaction on developing thymocytes likely generates qualitatively distinct signaling critical for the CD4/CD8 lineage choice. Future research aimed at dissecting TCR signaling, both quantitative and qualitative, in inducing Thpok in MHCIIsignaled thymocytes and Runx3 in MHCI- signaled thymocytes will fill the crucial gap in our understanding of the CD4 helper versus CD8 cytotoxic lineage choice. This information may be useful in identifying pathways that can be targeted in conditions of autoimmunity or inflammation involving the two types of mature T cells.
References
2. Rothenberg EV. Programming for T-lymphocyte fates: modularity and mechanisms. Genes & development. 2019 Sep 1;33(17-18):1117-35.
3. Inglesfield S, Cosway EJ, Jenkinson WE, Anderson G. Rethinking thymic tolerance: lessons from mice. Trends in Immunology. 2019 Apr 1;40(4):279-91.
4. Yi J, Kawabe T, Sprent J. New insights on T-cell selftolerance. Current Opinion in Immunology. 2020 Apr 1;63:14-20.
5. Liu X, Bosselut R. Duration of TCR signaling controls CD4-CD8 lineage differentiation in vivo. Nature Immunology. 2004 Mar;5(3):280-8.
6. Irie HY, Mong MS, Itano A, Crooks MC, Littman DR, Burakoff SJ, Robey E. The cytoplasmic domain of CD8ß regulates Lck kinase activation and CD8 T cell development. The Journal of Immunology. 1998 Jul 1;161(1):183-91.
7. Legname G, Seddon B, Lovatt M, Tomlinson P, Sarner N, Tolaini M, et al. Inducible expression of a p56Lck transgene reveals a central role for Lck in the differentiation of CD4 SP thymocytes. Immunity. 2000 May 1;12(5):537- 46.
8. Hernández-Hoyos G, Sohn SJ, Rothenberg EV, Alberola-Ila J. Lck activity controls CD4/CD8 T cell lineage commitment. Immunity. 2000 Mar 1;12(3):313-22.
9. Sohn SJ, Forbush KA, Pan XC, Perlmutter RM. Activated p56lck directs maturation of both CD4 and CD8 single-positive thymocytes. The journal of immunology. 2001 Feb 15;166(4):2209-17.
10. Li XL, Teng MK, Reinherz EL, Wang JH. Strict major histocompatibility complex molecule class-specific binding by co-receptors enforces MHC-restricted aß TCR recognition during T lineage subset commitment. Frontiers in Immunology. 2013 Nov 22;4:383.
11. Kisielow P. How does the immune system learn to distinguish between good and evil? The first definitive studies of T cell central tolerance and positive selection. Immunogenetics. 2019 Aug 15:1-6.
12. Sarafova SD, Erman B, Yu Q, Van Laethem F, Guinter T, Sharrow SO, et al. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity. 2005 Jul 1;23(1):75-87.
13. Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4-versus CD8-lineage choice. Nature Reviews Immunology. 2008 Oct;8(10):788-801.
14. Erman B, Alag AS, Dahle O, van Laethem F, Sarafova SD, Guinter TI, et al. Coreceptor signal strength regulates positive selection but does not determine CD4/CD8 lineage choice in a physiologic in vivo model. The journal of Immunology. 2006 Nov 15;177(10):6613-25.
15. Park JH, Adoro S, Lucas PJ, Sarafova SD, Alag AS, Doan LL, et al. ‘Coreceptor tuning’: cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nature Immunology. 2007 Oct;8(10):1049-59.
16. Kimura MY, Pobezinsky LA, Guinter TI, Thomas J, Adams A, Park JH, Tai X, Singer A. IL-7 signaling must be intermittent, not continuous, during CD8+ T cell homeostasis to promote cell survival instead of cell death. Nature immunology. 2013 Feb;14(2):143-51.
17. Park JH, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nature Immunology. 2010 Mar;11(3):257-64.
18. Taniuchi I. CD4 helper and CD8 cytotoxic T cell differentiation. Annual Review of Immunology. 2018 Apr 26;36:579-601.
19. Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, et al. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nature Immunology. 2008 Oct;9(10):1113-21.
20. He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005 Feb;433(7028):826-33.
21. Dave VP, Allman D, Keefe R, Hardy RR, Kappes DJ. HD mice: a novel mouse mutant with a specific defect in the generation of CD4+ T cells. Proceedings of the National Academy of Sciences. 1998 Jul 7;95(14):8187-92.
22. Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nature Immunology. 2005 Apr;6(4):373-81.
23. Sato T, Ohno SI, Hayashi T, Sato C, Kohu K, Satake M, et al. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity. 2005 Mar 1;22(3):317-28.
24. Kohu K, Sato T, Ohno SI, Hayashi K, Uchino R, Abe N, et al. Overexpression of the Runx3 transcription factor increases the proportion of mature thymocytes of the CD8 single-positive lineage. The Journal of Immunology. 2005 Mar 1;174(5):2627-36.
25. Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, Bernstein Y, Goldenberg D, Brenner O, Berke G, Levanon D. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proceedings of the National Academy of Sciences. 2003 Jun 24;100(13):7731- 6.
26. Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002 Nov 27;111(5):621- 33.
27. Sinclair C, Ono M, Seddon B. A Zap70-dependent feedback circuit is essential for efficient selection of CD4 lineage thymocytes. Immunology and Cell Biology. 2015 Apr;93(4):406-16.
28. Sinclair C, Seddon B. Overlapping and asymmetric functions of TCR signaling during thymic selection of CD4 and CD8 lineages. The Journal of Immunology. 2014 Jun 1;192(11):5151-9.
29. Saini M, Sinclair C, Marshall D, Tolaini M, Sakaguchi S, Seddon B. Regulation of Zap70 expression during thymocyte development enables temporal separation of CD4 and CD8 repertoire selection at different signaling thresholds. Science signaling. 2010 Mar 23;3(114):ra23.
30. Jenkinson SR, Intlekofer AM, Sun G, Feigenbaum L, Reiner SL, Bosselut R. Expression of the transcription factor cKrox in peripheral CD8 T cells reveals substantial postthymic plasticity in CD4-CD8 lineage differentiation. The Journal of Experimental Medicine. 2007 Feb 19;204(2):267-72.
31. Zeidan N, Damen H, Roy DC, Dave VP. Critical role for TCR signal strength and MHC specificity in ThPOKInduced CD4 helper lineage choice. The Journal of Immunology. 2019 Jun 1;202(11):3211-25.
32. Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, et al. Runx3 regulates integrin aE/CD103 and CD4 expression during development of CD4-/ CD8+ T cells. The Journal of Immunology. 2005 Aug 1;175(3):1694-705.
33. Issuree PD, Day K, Au C, Raviram R, Zappile P, Skok JA, et al. Stage-specific epigenetic regulation of CD4 expression by coordinated enhancer elements during T cell development. Nature communications. 2018 Sep 5;9(1):1-3.
34. Issuree PD, Ng CP, Littman DR. Heritable gene regulation in the CD4: CD8 T cell lineage choice. Frontiers in Immunology. 2017 Mar 22;8:291.
35. Tanaka H, Naito T, Muroi S, Seo W, Chihara R, Miyamoto C, et al. Epigenetic Thpok silencing limits the time window to choose CD4+ helper-lineage fate in the thymus. The EMBO Journal. 2013 Apr 17;32(8):1183-94.
36. Xing S, Shao P, Li F, Zhao X, Seo W, Wheat JC, et al. Tle corepressors are differentially partitioned to instruct CD8+ T cell lineage choice and identity. Journal of Experimental Medicine. 2018 Aug 6;215(8):2211-26.
37. Sakaguchi S, Hainberger D, Tizian C, Tanaka H, Okuda T, Taniuchi I, Ellmeier W. MAZR and Runx factors synergistically repress ThPOK during CD8+ T cell lineage development. The Journal of Immunology. 2015 Sep 15;195(6):2879-87.
38. Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, et al. Repression of the transcription factor Th- POK by Runx complexes in cytotoxic T cell development. Science. 2008 Feb 8;319(5864):822-5.
39. Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nature Immunology. 2008 Oct;9(10):1131-9.
40. Xing S, Li F, Zeng Z, Zhao Y, Yu S, Shan Q, et al. Tcf1 and Lef1 transcription factors establish CD8+ T cell identity through intrinsic HDAC activity. Nature Immunology. 2016 Jun;17(6):695-703.
41. Kakugawa K, Kojo S, Tanaka H, Seo W, Endo TA, Kitagawa Y, et al. Essential roles of SATB1 in specifying T lymphocyte subsets. Cell Reports. 2017 May 9;19(6):1176- 88.
42. Kojo S, Tanaka H, Endo TA, Muroi S, Liu Y, Seo W, et al. Priming of lineage-specifying genes by Bcl11b is required for lineage choice in post-selection thymocytes. Nature Communications. 2017 Sep 26;8(1):1-4.
43. Kojo S, Yasmin N, Muroi S, Tenno M, Taniuchi I. Runx-dependent and silencer-independent repression of a maturation enhancer in the Cd4 gene. Nature Communications. 2018 Sep 5;9(1):1-1.
44. He X, Park K, Wang H, He X, Zhang Y, Hua X, et al. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008 Mar 14;28(3):346-58.
45. Muroi S, Tanaka H, Miyamoto C, Taniuchi I. Cutting edge: fine-tuning of Thpok gene activation by an enhancer in close proximity to its own silencer. The Journal of Immunology. 2013 Feb 15;190(4):1397-401.
46. Wang L, Carr T, Xiong Y, Wildt KF, Zhu J, Feigenbaum L, Bendelac A, Bosselut R. The sequential activity of Gata3 and Thpok is required for the differentiation of CD1d-restricted CD4+ NKT cells. European Journal of Immunology. 2010 Sep;40(9):2385-90.
47. Gimferrer I, Hu T, Simmons A, Wang C, Souabni A, Busslinger M, et al. Regulation of GATA-3 expression during CD4 lineage differentiation. The Journal of Immunology. 2011 Apr 1;186(7):3892-8.
48. Xiong Y, Castro E, Yagi R, Zhu J, Lesourne R, Love PE, et al. Thpok-independent repression of R unx3 by G ata3 during CD 4+ T-cell differentiation in the thymus. European Journal of Immunology. 2013 Apr;43(4):918- 28.
49. Tsagaratou A, González-Avalos E, Rautio S, Scott- Browne JP, Togher S, Pastor WA, et al. TET proteins regulate the lineage specification and TCR-mediated expansion of i NKT cells. Nature Immunology. 2017 Jan;18(1):45-53.
50. Tsagaratou A, Lio CW, Yue X, Rao A. TET methylcytosine oxidases in T cell and B cell development and function. Frontiers in immunology. 2017 Mar 31;8:220.
51. Luckey MA, Kimura MY, Waickman AT, Feigenbaum L, Singer A, Park JH. The transcription factor ThPOK suppresses Runx3 and imposes CD4+ lineage fate by inducing the SOCS suppressors of cytokine signaling. Nature Immunology. 2014 Jul;15(7):638-45.
52. Ilangumaran S, Bobbala D, Ramanathan S. SOCS1: regulator of T cells in autoimmunity and cancer. InEmerging Concepts Targeting Immune Checkpoints in Cancer and Autoimmunity 2017 (pp. 159-189).
53. Hwang S, Lee C, Park K, Oh S, Jeon S, Kang B, et al. Twist2 promotes CD8+ T-cell differentiation by repressing ThPOK expression. Cell Death & Differentiation. 2020 May 18:1-2.
54. Nowyhed HN, Huynh TR, Blatchley A, Wu R, Thomas GD, Hedrick CC. The nuclear receptor nr4a1 controls CD8 T cell development through transcriptional suppression of runx3. Scientific Reports. 2015 Mar 12;5(1):1-9