Abstract
In this article, we introduced a screening of anti-fibrotic drugs focused on new genes. More precisely, we screened and cloned 127 new genes, reporting on a potential target gene and two promising drugs for fibrosis. Among 127 genes, hepatitis C virus nonstructural protein 5A transactivated protein 9 (NS5ATP9), which expression is significantly upregulated by tenofovir disoproxil fumarate (TDF)/tenofovir alafenamide fumarate (TAF), suppresses hepatic stellate cells (HSCs) and HFL1 cells (lung fibroblasts) activation. Therefore, we reported NS5ATP9 as a potential therapeutic target, and TDF/TAF as a new promising therapeutic strategy in fibrosis. These results elucidate mechanisms of disease and translate molecular techniques into clinical treatment.
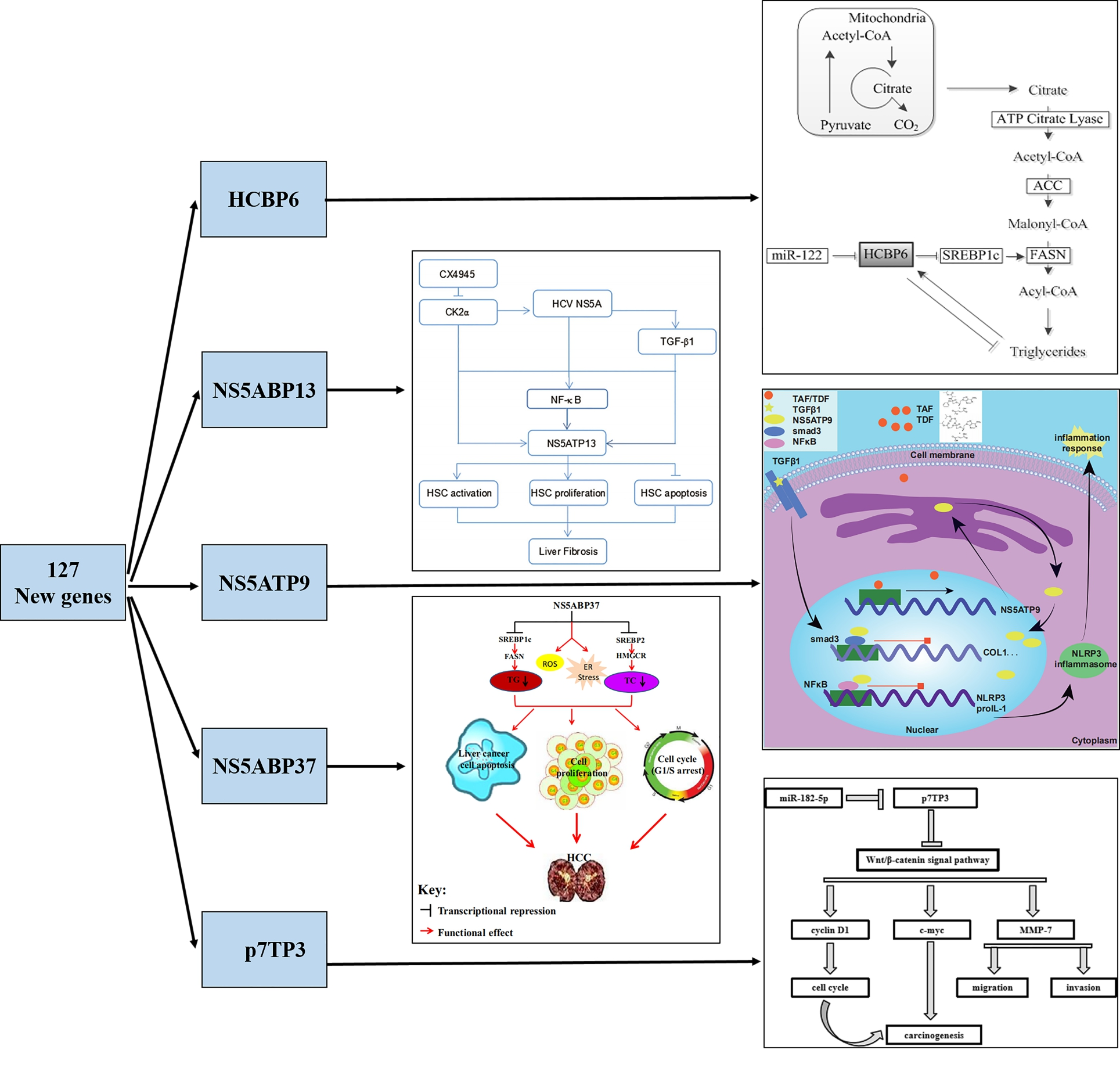
Discovery of 127 New Genes and the Main Achievements
By the suppression of subtractive hybridization (SSH) and yeast-two hybrid system, 127 new genes were screened, cloned, and registered at GenBank (Table 1) [1,2]. These new genes which were found in the liver have been demonstrated to be closely related to liver diseases such as viral hepatitis, liver fibrosis, fatty liver, and hepatocellular carcinoma (HCC) (Figure 1).
| Serial number | Gene name | Registration number (GenBank) |
|---|---|---|
| 1 | NS5ATP1 | AF529362 |
| 2 | NS5ATP2 | AF529363 |
| 3 | NS5ATP3 | AF529364 |
| 4 | NS5ATP4 | AF529365 |
| 5 | NS5ATP5 | AF529366 |
| 6 | NS5ATP6 | AF529367 |
| 7 | NS5ATP7 | AF529368 |
| 8 | NS5ATP8 | AF529369 |
| 9 | NS5ATP9 | AF529370 |
| 10 | NS5ATP10 | AK000514 |
| 11 | NS5ATP11 | AK091427 |
| 12 | DNAPTP1TPA | DQ414820 |
| 13 | NS5ATP13 | AY820769 |
| 14 | NS5BBP1 | BC020596 |
| 15 | MBP1 | DQ307498 |
| 16 | XTP1 | AF488828 |
| 17 | XTP2 | AF488829 |
| 18 | XTP3 | AF490252 |
| 19 | XTP4 | AF490253 |
| 20 | XTP5 | AF490254 |
| 21 | XTP6 | AF490255 |
| 22 | XTP7 | AF490256 |
| 23 | XTP8 | AF490257 |
| 24 | XTP9 | AF490258 |
| 25 | XTP10 | NM_001326303 |
| 26 | X-30 | AY280722 |
| 27 | C1 | AY555145 |
| 28 | C2 | AF530058 |
| 29 | C12 | AF529371 |
| 30 | E2BP1 | AY459290 |
| 31 | E2BP2 | AF529373 |
| 32 | E2BP3 | DQ294736 |
| 33 | E2BP4 | AF189768 |
| 34 | EBP1 | AF529372 |
| 35 | EBP2 | AF529373 |
| 36 | EBP3 | AF530058 |
| 37 | EBP4 | AY134474 |
| 38 | EBP19 | AF529373 |
| 39 | EBP36 | AY189820 |
| 40 | HCBP1 | AF359506 |
| 41 | HCBP6 | AY032594 |
| 42 | HCBP12 | AF395068 |
| 43 | HCTP4 | AY734680 |
| 44 | NS3BP | AF435951 |
| 45 | NS3TP1 | AY116969 |
| 46 | NS3TP2 | AY116970 |
| 47 | NS3TP6 | XM_017004595 |
| 48 | PreS1BP1 | AY535000 |
| 49 | PS2BP1 | AF497566 |
| 50 | SBP1 | AY281252 |
| 51 | TAHCCP1 | AY038359 |
| 52 | TAHCCP2 | AY039043 |
| 53 | TTP1 | AF407672 |
| 54 | XBP-1 | AF529374 |
| 55 | LRRP1 | AY358788 |
| 56 | PS1TP1 | AY646229 |
| 57 | PS1TP2 | AY426673 |
| 58 | PS1TP3 | AY426674 |
| 59 | PS1TP4 | AY427952 |
| 60 | PS1TP5TP1 | ABF61801 |
| 61 | PS1TP6 | AY444749 |
| 62 | PS2TP1 | AY561706 |
| 63 | PS2TP2 | AY561707 |
| 64 | PS2TP3 | AY561704 |
| 65 | PS2TP4 | AY561705 |
| 66 | CSTP1 | AY553877 |
| 67 | PS1TP3BP1 | DQ910907 |
| 68 | NS5ATP13TP1 | AY459295 |
| 69 | NS5ATP13TP2 | AY459296 |
| 70 | HBeAgTP | AY423624 |
| 71 | PFAAP1 | AF530059 |
| 72 | PFAAP2 | AF530060 |
| 73 | PFAAP3 | AF530061 |
| 74 | PFAAP4 | AF530062 |
| 75 | PFAAP5 | AF530063 |
| 76 | FBP2 | AY553876 |
| 77 | FBP1 | AY553875 |
| 78 | NS5ATP5BP1 | AY459291 |
| 79 | HCTP4BP | AY390431 |
| 80 | NS5ATP1BP16 | AY390430 |
| 81 | NS5ABP37 | AF543840 |
| 82 | XTP3TPB | AY453410 |
| 83 | XTP3TPA | AY453409 |
| 84 | DNAPTP1 | AY450389 |
| 85 | DNAPTP2 | AY450390 |
| 86 | DNAPTP3 | AY450391 |
| 87 | DNAPTP4 | AY450392 |
| 88 | DNAPTP5 | AY450393 |
| 89 | DNAPTP6 | AY450394 |
| 90 | PPS22-1 | AY498718 |
| 91 | NS3TP2TP | AY425618 |
| 92 | NS5ATP6TP1 | AY339614 |
| 93 | NS5ATP6TP2 | AY339615 |
| 94 | HuALR | AF146394 |
| 95 | P7TP3 | AY820138 |
| 96 | P7TP2 | AY819648 |
| 97 | AsTP3 | AY744367 |
| 98 | AsTP2 | AY744366 |
| 99 | FTP2 | AY740522 |
| 100 | AsTP | AY720898 |
| 101 | AsTP1 | AY605064 |
| 102 | XTP13 | AY631401 |
| 103 | NS5ATP4A | DQ908899 |
| 104 | NS5ATP4ABP1 | DQ630520 |
| 105 | PS1TP5 | AY427953 |
| 106 | XTP3TPATP1 | DQ457058 |
| 107 | FTP1 | AY605045 |
| 108 | XTP12 | AY598792 |
| 109 | PS1TP5BP1 | DQ471327 |
| 110 | P7TP1 | AY596776 |
| 111 | NS3TP6BP2 | AC097504 |
| 112 | NS3TP6BP3 | AC023785 |
| 113 | TTG1 | DQ323046 |
| 114 | XTP11 | AY740520 |
| 115 | DNAPTP1BP1 | DQ414819 |
| 116 | DNAPTP1TP | DQ451688 |
| 117 | HBEBP2BPA | DQ499597 |
| 118 | HBEBP2BPB | DQ499598 |
| 119 | HBEBP2BPC | DQ499599 |
| 120 | NS2TP | AY605046 |
| 121 | NS4ATP1 | AY740521 |
| 122 | NS4ATP2 | AY846876 |
| 123 | TTG1A | DQ529299 |
| 124 | PS1TP2BP1 | DQ787424 |
| 125 | HCBP12BPA | DQ499468 |
| 126 | XTP3TPATP2 | DQ457059 |
| 127 | NS3TP6BP1 | AC124014 |
For several decades, our group was committed to studying of these 127 new genes, providing a new research perspective for liver diseases. Hepatitis C virus core protein-binding protein 6 (HCBP6) upregulates sterol regulatory element-binding protein 1c (SREBP1c) expression by binding to the C/EBPβ-binding site in the SREBP1c promoter [3] and then modulating intracellular triglyceride homeostasis [4]. Hepatitis C virus nonstructural protein 5A trans-activated protein 6 (NS5ATP6) regulates the intracellular triglyceride level via fibroblast growth factor 21 (FGF21), and independently of sirtuin1 (SIRT1) and SREBP1 [5]. HCV promotes the profibrogenic effect of HCV NS5A-transactivated protein 13 (NS5ATP13), by transforming growth factor β1/Sekelsky mothers against decapentaplegic homolog 3 (TGFβ1/Smad3) and nuclear factor κB (NF-κB) signal pathways. Moreover, as a pro-fibrogenic factor, NS5ATP13 expression is down-regulated by CX-4945, a CK2 specific inhibitor [6]. Besides, NS5ATP13 promotes the proliferation and migration of HepG2 cells (human hepatoblastoma HepG2 cell line). Also, oxymatrine (OMT) may inhibit liver cancer progression by downregulating NS5ATP13 expression [7]. Hepatitis C virus nonstructural protein 5A-associated binding protein 37 (NS5ABP37) inhibits cancer cell proliferation and promotes its apoptosis, by altering SREBP-dependent lipogenesis and cholesterogenesis and inducing oxidative stress and endoplasmic reticulum stress [8]. In HCC, hepatitis B virus X Ag-transactivated protein 8 (XTP8) acts as a valuable prognostic predictor by forming a positive feedback loop with FOXM1 oncogene [9]. Hepatitis C virus p7 trans-regulated protein 3 (p7TP3), the direct target gene of miR-182-5p, inhibits HCC by suppressing migration, invasion, adhesion, proliferation and cell cycle progression of liver cancer cell via Wnt/β- catenin signaling pathway, which suggests that p7TP3 might be a new promising tumor suppressor [10]. HBX protein trans-activate gene (XTP4) suppresses apoptosis of HepG2 by up-regulating Bcl-2 and Bax expression [11], and promotes the migration and invasion of HepG2 via regulation of epithelial-mesenchymal transition (EMT) related molecules E-cadherin and N-cadherin [12]. HBV PS1 trans-activator protein 2 (PS1TP2) inhibits apoptosis of HepG2 via the mitochondrial pathway, and promotes proliferation via adenosine 5-monophosphate-activated protein kinase (AMPK) pathway [13]. Besides, NS5ATP9 is a new gene that has been widely recognized in various fields over recent years.
For several decades, our group was committed to studying of these 127 new genes, providing a new research perspective for liver diseases. Hepatitis C virus core protein-binding protein 6 (HCBP6) upregulates sterol regulatory element-binding protein 1c (SREBP1c) expression by binding to the C/EBPβ-binding site in the SREBP1c promoter [3] and then modulating intracellular triglyceride homeostasis [4]. Hepatitis C virus nonstructural protein 5A trans-activated protein 6 (NS5ATP6) regulates the intracellular triglyceride level via fibroblast growth factor 21 (FGF21), and independently of sirtuin1 (SIRT1) and SREBP1 [5]. HCV promotes the profibrogenic effect of HCV NS5A-transactivated protein 13 (NS5ATP13), by transforming growth factor β1/Sekelsky mothers against decapentaplegic homolog 3 (TGFβ1/Smad3) and nuclear factor κB (NF-κB) signal pathways. Moreover, as a pro-fibrogenic factor, NS5ATP13 expression is down-regulated by CX-4945, a CK2 specific inhibitor [6]. Besides, NS5ATP13 promotes the proliferation and migration of HepG2 cells (human hepatoblastoma HepG2 cell line). Also, oxymatrine (OMT) may inhibit liver cancer progression by downregulating NS5ATP13 expression [7]. Hepatitis C virus nonstructural protein 5A-associated binding protein 37 (NS5ABP37) inhibits cancer cell proliferation and promotes its apoptosis, by altering SREBP-dependent lipogenesis and cholesterogenesis and inducing oxidative stress and endoplasmic reticulum stress [8]. In HCC, hepatitis B virus X Ag-transactivated protein 8 (XTP8) acts as a valuable prognostic predictor by forming a positive feedback loop with FOXM1 oncogene [9]. Hepatitis C virus p7 trans-regulated protein 3 (p7TP3), the direct target gene of miR-182-5p, inhibits HCC by suppressing migration, invasion, adhesion, proliferation and cell cycle progression of liver cancer cell via Wnt/β- catenin signaling pathway, which suggests that p7TP3 might be a new promising tumor suppressor [10]. HBX protein trans-activate gene (XTP4) suppresses apoptosis of HepG2 by up-regulating Bcl-2 and Bax expression [11], and promotes the migration and invasion of HepG2 via regulation of epithelial-mesenchymal transition (EMT) related molecules E-cadherin and N-cadherin [12]. HBV PS1 trans-activator protein 2 (PS1TP2) inhibits apoptosis of HepG2 via the mitochondrial pathway, and promotes proliferation via adenosine 5-monophosphate-activated protein kinase (AMPK) pathway [13]. Besides, NS5ATP9 is a new gene that has been widely recognized in various fields over recent years.
NS5ATP9
NS5ATP9 genomic DNA, which is located on human chromosome 15q22.1, encodes a protein with 111 amino acid residues [14]. It is also known as KIAA0101, OEACT-1, P15PAF, L5, PCNA-associated factor (PAF), and is registered in GenBank under the AF529370 registration number. NS5ATP9 participates in many physiological functions, such as cartilage formation [15], DNA damage repair [16], cell cycle regulation [17], the maturation and development of hematopoietic stem/progenitor cells [18], and so on. In addition, different kinds of tumor development are associated with uncontrolled expression of NS5ATP9, including HCC [19], breast cancer [20], thyroid carcinoma [21], and non-small cell lung cancer [22].
Endogenous NS5ATP9 expression is overexpressed in CCl4-induced liver fibrosis mouse models and TGFβ1- treated hepatic stellate cells (HSCs) [23]. In LX2 cells (human HSC cell line), NS5ATP9 directly binds to Smad3 and inhibits its phosphorylation, which induces [24]. Besides, compared with wild type mice, NS5ATP9 the suppression of the TGFβ1/Smad3 signal pathway deficiency results in significantly higher levels of ECM deposition, indicating that NS5ATP9 attenuates liver fibrosis in vivo and in vitro [23]. In lung fibroblasts, NS5ATP9 suppresses its activation via the TGFβ1/ Smad3 signal pathway [25]. These studies confirmed that NS5ATP9 inhibits liver fibrosis and lung fibrosis.
Drugs Screening
Given that NS5ATP9 is a potential therapeutic target for liver fibrosis and lung fibrosis, drugs or small molecule compounds targeted at NS5ATP9 are expected to treat fibrosis. In primary vaginal epithelial cells, expression of NS5ATP9 in mRNA level is up-regulated after cells are stimulated by TDF for 1 or 7 days [26]. Therefore, we hypothesized that TDF and its pro-drug, TAF, may promote regression of fibrosis via up-regulated NS5ATP9.
In vivo, TAF inhibits both CCl4-induced liver fibrosis and bleomycin-induced pulmonary fibrosis, while TAF inhibits activation of HSCs and lung fibroblasts in vitro [23,25]. Previous studies have also shown that TDF/ TAF inhibit liver fibrosis by inhibiting TGFβ1/Smad3 and NF-κB/NLRP3 inflammasome signaling pathways activation and regulating the differentiation, activation, and proliferation of HSCs [23].
Consistent with previous findings [14], TDF/TAF upregulate NS5ATP9 expression both in the liver and in the lung. By using dual-luciferase reporter assays, we showed that NS5ATP9 promoter activity was upregulated by TDF and TAF. Therefore, TDF/TAF could prevent progression and promote the reversion of fibrosis by upregulating the expression of NS5ATP9.
Conclusions and Perspectives
In summary, our study proposed a novel role of TDF/ TAF in fibrosis progression through assembling TGFβ1/ Smad3 and NF-κB/NLRP3 inflammasome signaling pathways via upregulating the expression of NS5ATP9, thus defining NS5ATP9 as a potential therapeutic target and TDF/TAF as novel drugs for fibrosis.
Challenges
Fibrosis is defined as the accumulation of extracellular matrix (ECM) in specific organs. TDF/TAF inhibits liver fibrosis and lung fibrosis in mouse models. However, to elucidate the role of TDF/TAF in clinic, we asked the following questions: 1. Do the results from mouse experiments translate to human liver fibrosis and lung fibrosis? So results in a large, prospective, double-blind study are needed [27]. 2. Do TDF and TAF inhibit fibrosis in other organs or fibrosis due to other causes, such as bile duct ligation (BDL)-induced liver fibrosis [28]? 3. When used to treat different organ fibrosis, what is the optimal time and dosages of TDF/TAF? This means that the pharmacokinetics of TDF and TAF in liver fibrosis are indispensable [29]. 4. New insight into liver fibrosis therapy is that the intercellular crosstalk between HSCs and those “responded” cells (such as hepatic macrophages and natural killer/natural killer T cells) has been a critical event involved in HSC activation and fibrogenesis [30]. We propose that TDF and TAF inhibit NF-κB/NLRP3 inflammasome in mice [17]. However, how TDF and TAF affect the inflammation and immune system is not completely solved.
Opportunities
Compared with other novel treatment strategies, such as low-energy extracorporeal shock waves [31], hyperbranched lipoid-based lipid nanoparticles [32], acetyl-CoA carboxylase [33], as marketed drugs, adverse reactions to TDF/TAF can be effectively followed up with broad physician support, and an adequate number of patients. In addition, the cooperation of multiple clinical departments could fill the gap related to TDF/TAF in the treatment of fibrosis affecting other organs.
Acknowledgments
This study was supported by the National Key Research and Development Program of China (No. 2017YFC0908100/2017YFC0908104), the National Natural Science Foundation of China (No. 81670547), the Beijing Municipal Administration of Hospitals (XMLX201711 to JC), the Beijing Municipal Administration of Hospitals’ Ascent Plan (DFL20151701), and the National Science and Technology Major Project (No. 2015ZX10004801-001-002, No. 2017ZX10302201- 005-004 and No. 2017ZX10202202-005-008). Support was also provided by the Program of Beijing Advanced Innovation Center for Big Data-Based Precision Medicine and the Beijing Key Laboratory of Emerging Infectious Diseases, Beijing, China.
References
2. Bai GQ, Liu Y, Cheng J, Zhang SL, Yue YF, Huang YP, Zhang LY. Transactivating effect of complete S protein of hepatitis B virus and cloning of genes transactivated by complete S protein using suppression subtractive hybridization technique. World Journal of Gastroenterology: WJG. 2005 Jul 7;11(25):3893.
3. Yang X, Han M, Liu S, Yuan X, Liu X, Feng S, Zhou L, Li Y, Lu H, Cheng J, Lin S. HCBP6 upregulates human SREBP1c expression by binding to C/EBPβ-binding site in the SREBP1c promoter. BMB reports. 2018 Jan;51(1):33.
4. Gao LL, Li M, Wang Q, Liu SA, Zhang JQ, Cheng J. HCBP6 modulates triglyceride homeostasis in hepatocytes via the SREBP1c/FASN pathway. Journal of cellular biochemistry. 2015 Oct;116(10):2375-84.
5. Li Z, Feng S, Zhou L, Liu S, Cheng J. NS5ATP6 modulates intracellular triglyceride content through FGF21 and independently of SIRT1 and SREBP1. Biochemical and biophysical research communications. 2016 Jun 17;475(1):133-9.
6. Li Y, Liu S, Han M, Lu H, Wang Q, Zhang Y, et al. NS5ATP13 Promotes Liver Fibrogenesis Via Activation of Hepatic Stellate Cells. Journal of cellular biochemistry. 2017;118(8):2463-73. [PubMed: 28133777]
7. Yin Yue, Han Ming, Zhu Xiaoning, Zhang Yurong, Cheng Jun, Wang Jing. The mechanism of oxymatrine regulating the proliferation and apoptosis of human hepatoblastoma HepG2 cell line by NS5ATP13. Chinese Journal of Hepatology (Electronic Edition). 2019 (2): 1.
8. Feng S, Han M, Zhou L, Wang Q, Li Z, Li Y, Lu H, Liu T, Ma Y, Liu S, Cheng J. NS 5 ABP 37 inhibits liver cancer by impeding lipogenesis and cholesterogenesis. Cancer science. 2017 Jan;108(1):12-22.
9. Han M, Lu H, Han K, Yuan X, Liu S, Wang Y, et al. XTP8 promotes hepatocellular carcinoma growth by forming a positive feedback loop with FOXM1 oncogene. Biochemical and biophysical research communications. 2019 Jul 30;515(3):455-61.
10. Zhao J, Wang Y, Han M, Lu H, Chen X, Liu S, Yuan X, Han K, Liang P, Cheng J. P7TP3 inhibits tumor development, migration, invasion and adhesion of liver cancer via Wnt/β-catenin signal pathway. Cancer science. 2019 Nov 20.
11. Shi C, Zhang M, Wang L, Liu S, Liang P, Wu J, et al. HBX protein trans-activate gene XTP4 suppresses HepG2 cells apoptosis. Basic & clinical medicine. 2015;35(12):1591-95.
12. Deng X, Han M, Liu S, Cheng J, Liang Y. HBX protein trans-activate gene XTP4 promotes the migration and invasion of hepatocellular carcinoma cell line]. Basic & clinical medicine. 2018;38(12):1718-1723.
13. Ju W, Li Q , Han M, Liu S, Wu J, Cheng J, et al. Effect of HBV PS1 trans-activator protein-2 gene on proliferation and apoptosis of HepG2 cells. Chongqing medicine. 2019;48(12):2001-2005.
14. Li K, Ma Q, Shi L, Dang C, Hong Y, Wang Q, Li Y, Fan W, Zhang L, Cheng J. NS5ATP9 gene regulated by NF-κB signal pathway. Archives of biochemistry and biophysics. 2008 Nov 1;479(1):15-9.
15. Sang Y, Zang W, Yan Y, Liu Y, Fu Q, Wang K, Chen Y, Qi N. Study of differential effects of TGF-beta3/BMP2 on chondrogenesis in MSC cells by gene microarray data analysis. Molecular and cellular biochemistry. 2014 Jan 1;385(1-2):191-8.
16. Li G, Luna C, Gonzalez P. miR-183 inhibits UVinduced DNA damage repair in human trabecular meshwork cells by targeting of KIAA0101. Investigative ophthalmology & visual science. 2016 Apr 1;57(4):2178- 86.
17. Karg E, Smets M, Ryan J, Forné I, Qin W, Mulholland CB, Kalideris G, Imhof A, Bultmann S, Leonhardt H. Ubiquitome analysis reveals PCNA-associated factor 15 (PAF15) as a specific ubiquitination target of UHRF1 in embryonic stem cells. Journal of molecular biology. 2017 Dec 8;429(24):3814-24.
18. Wang X, Jung YS, Jun S, Lee S, Wang W, Schneider A, Oh YS, Lin SH, Park BJ, Chen J, Keyomarsi K. PAFWnt signaling-induced cell plasticity is required for maintenance of breast cancer cell stemness. Nature communications. 2016 Feb 4;7:10633.
19. Abdelgawad IA, Radwan NH, Hassanein HR. KIAA0101 mRNA expression in the peripheral blood of hepatocellular carcinoma patients: Association with some clinicopathological features. Clinical biochemistry. 2016 Jul 1;49(10-11):787-91.
20. Zhang HF, Alshareef A, Wu C, Jiao JW, Sorensen PH, Lai R, Xu LY, Li EM. miR-200b induces cell cycle arrest and represses cell growth in esophageal squamous cell carcinoma. Carcinogenesis. 2016 Sep 1;37(9):858-69.
21. Mizutani K, Onda M, Asaka S, Akaishi J, Miyamoto S, Yoshida A, Nagahama M, Ito K, Emi M. Overexpressed in anaplastic thyroid carcinoma-1 (OEATC-1) as a novel gene responsible for anaplastic thyroid carcinoma. Cancer. 2005 May 1;103(9):1785-90.
22. Kim MJ, Xia B, Suh HN, Lee SH, Jun S, Lien EM, Zhang J, Chen K, Park JI. Paf-myc-controlled cell stemness is required for intestinal regeneration and tumorigenesis. Developmental cell. 2018 Mar 12;44(5):582-96.
23. Zhao J, Han M, Zhou L, Liang P, Wang Y, Feng S, Lu H, Yuan X, Han K, Chen X, Liu S. TAF and TDF attenuate liver fibrosis through NS5ATP9, TGFβ1/Smad3, and NF-κB/NLRP3 inflammasome signaling pathways. Hepatology International. 2020 Jan;14(1):145-60.
24. Zhang M, Zhang J, Liu S, Wang Q, Lin G, Qiu R, Quan M, Cheng J. NS5ATP9 suppresses activation of human hepatic stellate cells, possibly via inhibition of Smad3/ phosphorylated-Smad3 expression. Inflammation. 2015 Feb 1;38(1):278-89.
25. Li L, Zhao J, Zhou L, Chen J, Ma Y, Yu Y, Cheng J. Tenofovir alafenamide fumarate attenuates bleomycininduced pulmonary fibrosis by upregulating the NS5ATP9 and TGF-β1/Smad3 signaling pathway. Respiratory research. 2019 Dec;20(1):163.
26. Hladik F, Burgener A, Ballweber L, Gottardo R, Vojtech L, Fourati S, Dai JY, Cameron MJ, Strobl J, Hughes SM, Hoesley C. Mucosal effects of tenofovir 1% gel. Elife. 2015 Feb 3;4:e04525.
27. Garcia-Tsao G, Bosch J, Kayali Z, Harrison SA, Abdelmalek MF, Lawitz E, Satapathy SK, Ghabril M, Shiffman ML, Younes ZH, Thuluvath PJ. Randomized Placebo-Controlled Trial of Emricasan in Non-alcoholic Steatohepatitis (NASH) Cirrhosis with Severe Portal Hypertension. Journal of Hepatology. 2019 Dec 21.
28. Baghdasaryan A, Claudel T, Kosters A, Gumhold J, Silbert D, Thüringer A, Leski K, Fickert P, Karpen SJ, Trauner M. Curcumin improves sclerosing cholangitis in Mdr2−/− mice by inhibition of cholangiocyte inflammatory response and portal myofibroblast proliferation. Gut. 2010 Apr 1;59(4):521-30.
29. Almomen A, Maher HM, Alzoman NZ, Shehata SM, Al-taweel SM, Alanazi AA. Development and validation of UPLC-MS/MS method for studying the pharmacokinetic interaction of dasabuvir and tamoxifen, 4-hydroxytamoxifen in Wistar rats. Scientific Reports. 2020;10.
30. Cai X, Wang J, Wang J, Zhou Q, Yang B, He Q, Weng Q. Intercellular crosstalk of hepatic stellate cells in liver fibrosis: new insights into therapy. Pharmacological Research. 2020 Feb 21:104720.
31. Ujiie N, Nakano T, Yamada M, Sato C, Nakanishi C, Fujishima F, Ito K, Shindo T, Shimokawa H, Kamei T. Low-energy extracorporeal shock wave therapy for a model of liver cirrhosis ameliorates liver fibrosis and liver function. Scientific Reports. 2020 Feb 12;10(1):1-7.
32. Qiao JB, Fan QQ, Zhang CL, Lee J, Byun J, Xing L, Gao XD, Oh YK, Jiang HL. Hyperbranched lipoid-based lipid nanoparticles for bidirectional regulation of collagen accumulation in liver fibrosis. Journal of Controlled Release. 2020 Mar 2.
33. Matsumoto M, Yashiro H, Ogino H, Aoyama K, Nambu T, Nakamura S, Nishida M, Wang X, Erion DM, Kaneko M. Acetyl-CoA carboxylase 1 and 2 inhibition ameliorates steatosis and hepatic fibrosis in a MC4R knockout murine model of nonalcoholic steatohepatitis. PloS one. 2020 Jan 28;15(1):e0228212.