Abstract
Renalase, a newly discovered flavin-adenine dinucleotide-dependent amine oxidase released by the kidney, circulates in the blood and has an impact on heart function and systemic blood pressure. We conducted the research in Babylon from February to July of 2016. We collected 50 blood samples from patients with end stage renal disease admitted to Merjan Teaching Hospital in Babylon Province, Iraq. In addition to the control group (50 samples) of healthy people aged 11 to 80, all patients and control groups were from the same ethnic group (Arab). The current study's findings revealed the mean variation of renalase level (pg/ml) across patient and control groups. Renalase levels were much higher (P <0.05) in the serum of people with end-stage renal disease (184.21± 4.97 pg/ml) than in the healthy group (44.06 ±7.21 pg/ml). The findings showed that people with end-stage renal failure were more likely to have GG, GA, and AA in the renalase (RNLS) gene (18%, 20%, and 62%) than people in the control group (19.3%, 22%, and 58.7%). In the Iraqi population, the Renalase (rs2576178) G>A mutation was not related with vulnerability to end-stage renal failure.
Keywords
Renalase, rs2576178, Polymorphisms, End stage renal disease, Kidney
Introduction
One flavoprotein enzyme that helps in catecholamine metabolism is renalase, which is another name for monoaminooxidase-C (MAO-C) [1]. With the help of nicotinamide adenine dinucleotide (NADH), superoxide breaks down catecholamines and other chemicals that have similar structures [2]. The renalase enzyme protein exists in a minimum of four different forms. While two of these, h-renalase 1 and 2, retain their original amino acid domains, the third and fourth, h-renalase 2, have shorter domains [3].
Unlike other amine oxidases, such as flavin adenine dinucleotide (FAD), the kidneys primarily produce and then release renalase into the bloodstream. It plays a role in the metabolism of catecholamines in circulation [4]. Pargyline and clorgyline, two well-known MAO inhibitors, have no effect on renalase activity [5].
Patients with chronic renal disease had decreased renalase concentrations, which may contribute to the onset of hypertension and its associated complications [2]. Researchers who looked at uremia in rats that had their kidneys removed confirmed that people with end-stage renal disease (ESRD) have a significant renalase deficiency [6]. This deficit increases the risk of cardiovascular disease by causing abnormal catecholamine breakdown and undue stress on the sympathetic nervous system [7].
Individuals with renal failure struggle to break down catecholamines, leading to elevated blood concentrations and an overactive sympathetic nervous system due to impaired renalase function. Guo et al. [8]. found that cardiovascular disease risk increases with sympathetic nervous system activity levels that are high. According to Malyszko et al. [9], the renalase gene (RNLS) is located on chromosome 10 (q23.33), contains 342 amino acids, and has a molecular weight of 37.8 kDa. The gene consists of 311,000 base pairs (bp) and 13 exons. Researchers have found multiple single nucleotide polymorphisms (SNPs) in the RNLS gene, indicating its high degree of polymorphism [10]. Most recently, researchers found that three RNLS gene SNPs (rs2296545, rs2576178, and rs10887800) were associated with several diseases. The rs2296545 (C/G) mutation is in the second exon of RNLS. It changes the Asp37Glu amino-acid substitution, which in turn changes how this gene works [11]. Intron 6 contains the polymorphism rs10887800, while the 5-flanking region contains the polymorphism rs2576178 (G/A). These SNPs may affect the regulation and gene expression of RNLS [12].
The aim of this study was to investigate the potential relationship between two polymorphisms in the renalase gene and hypertension in patients with ESRD.
Methods
Sampling
Between February 2016 and July 2016, researchers from the University of Babylon's Biotechnology and Genetic Engineering Laboratory, Department of Biology, College of Science, conducted a case-control study. Blood samples from patients undergoing dialysis at Iraq's Merjan Teaching Hospital in Babylon Province was collected.
This experiment made use of one hundred blood samples. We studied people with ESRD and 50 healthy individuals. The Merjan Teaching Hospital Dialysis Center had patients hospitalized for periods spanning eleven years to eighty years. Conversely, they matched the chronic kidney disease (CKD) and ESRD groups. A licensed physician examined each subject. People with hepatitis were not included.
Renalase quantity assay by ELISA
Elascience, a Chinese company, supplied the specific kit (ELISA) that was used to measure the level of human renalase.
DNA extraction
Using the Favergen Extraction and Purification Kit (Taiwan), genomic DNA was extracted and purified from whole blood cells.
Genotypic identification using RFLP- PCR amplification
We amplified the following DNA regions with specific primers. We detected polymorphisms in the renaelse gene using one primer (rs2576178) from Bioneer, IDTDNA (USA). Both the forward and reverse primer sequences were 5-AGCAGAGAAGCTTAACCT-3 and 5-TTATTGCAAGTCGTAAC-3, respectively. For the polymerase chain reaction (PCR), we made the reaction volume with 20 microlitres of water, 1 microlitre of each of the reverse and forward primers, 12.5 microlitres of Green Master Mix, 3 microlitres of genomic DNA, and 2.5 microlitres of nuclease-free water. We used thermocycling to achieve amplification, which involved two minutes at 94°C, thirty-five cycles of five minutes at 94°C, one minute at 60°C, and one minute at 72°C, followed by a final expansion of five minutes. After electrophoresing the PCR products in a 1% agarose gel at 75 V for 1 hour, we observed them using ethidium bromide. We captured the photos using the Gel documentation framework. We performed the PCR-RFLP technique, following the Promega Company Protocol, after cutting the PCR product using MSPI restriction endonuclease. After MSPI digestion, we subjected the reaction to electrophoresis on 3% agarose gels at 75 V for 1 hour and set up a control at 75 V, 20 Am for 160 minutes. We subsequently stained the gels with ethidium bromide. We captured images using the gel documentation system (EBOXCX, UK).
Statistical analysis
All statistical analysis were done using the SPSS applied mathematics software system (25; SPSS Inc., Chicago , IL), and P- values of <0.05 were considered statistically significant.
Results
Serum renalase concentration in both genders of the study groups
Table 1 displays the renalase levels (ng/ml) for both the patient and control groups. In contrast to the healthy control group, whose serum renalase levels were 44.06 ± 7.21 ng/ml, the ESRD group's levels were significantly higher (184.21 ± 4.97 ng/ml) (P <0.05).
|
Groups
|
ESRD |
Control |
P=value |
|
Renalase Level (ng/ml) |
184.21 ± 4.97 |
44.06 ± .21 |
0.001* |
|
*P ≤ 0.05; SE: Standard Error; ESRD: End Stage Renal Disease |
|||
Genetic polymorphisms of renalase gene associated with renal diseases
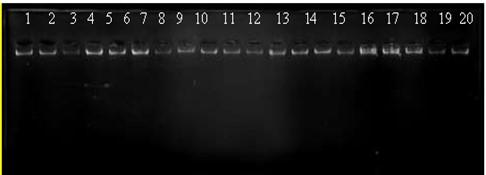
The genomic DNA (Figure 1) was extracted from the blood samples as a first step to amplify the target region of RNAL gene.
Figure 1. Genomic DNA electrophoresis pattern from renal patients and healthy control groups. The genomic DNA from blood samples (1-10 patients and 11-20 controls) is represented by lanes 1–20. The electrophoresis settings were 1% agarose, 75 V, 20 mA for 1 hour (10 µl in each well), and ethidium bromide was used for staining in the experiment.
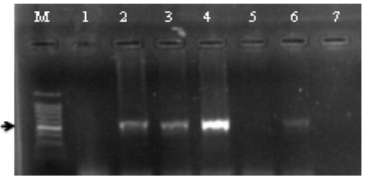
Figure 2. RNLS amplified product patterns of renal patients and healthy control groups as determined by agarose gel electrophoresis. The amplified products, which were a single band measuring 525 bp in size, are shown in Lanes 2-4 and 6, with M: standing for DNA size marker. The negative controls are located in Lanes 1 and 7. The conditions for electrophoresis and Ethidium bromide staining followed a 120-minute run at 75 V and 20 mA with a 1% agarose concentration.
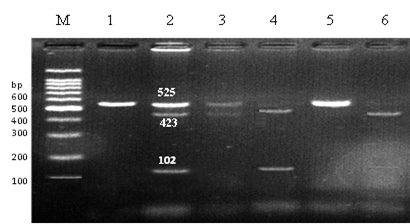
Figure 3 shows the results of cleaving the RNLS target sequence PCR products with the MspI (5' CCG 3') restriction enzyme, which allowed us to identify the rs2576178 SNP in the 5'-flanking region of the RNLS gene. Table 2 shows the data of the ESRD patient group, RNAL polymorphism frequencies were 62% and 18%, according to the PCR-RFLP findings compared to the healthy control group that possessed all three polymorphic alleles at rates of 19.3%, 58.7%, and 22%, respectively.
The digested products migrated into 3% agarose, 75 V, 20 mA for 120 min; 10 µl in each well; stained with ethidium bromide.
Figure 3. Electrophoresis patterns of allelotyping of the 5'-flanking region of the RNLS gene employing the MspI enzyme by PCR-RFLP technique in renal patients and healthy control groups. The DNA ladder ranges from 100 to 1100 base pairs. The first two lanes show a single band with a molecular size of 525 base pairs, while the second pair shows three bands with sizes of 525, 423, and 102 base pairs. The third, fourth, and sixth lanes show two bands with sizes of 423 and 102 base pairs, while the last pair represents a homozygous allele (GG).
|
Genotype |
ESRD No. (%) |
Control No. (%) |
Sig. |
OR (95%) ESRD |
|
AA a |
30 (62) |
27 (58.7%) |
1.00 |
|
|
AG |
12 (20) |
11 (22%) |
0.58 |
1.01 (0.38-2.68) |
|
GG |
8 (18) |
12 (19.3%) |
0.23 |
1.66 (0.50-4.69) |
|
Total number |
50 |
50 |
|
|
|
A allele |
72 |
65 |
0.28 |
1.09(0.60-1.97) |
|
G allele |
28 |
35 |
||
|
a reference; *P ≤ 0.05; OR= (95%CI); CKD: Chronic kidney disease; ESRD: End- stage renal disease |
||||
|
Variable |
(Mean ± SD) |
Genotype |
LSD |
|
ESRD |
134.97 ± 16.81 100.88 ± 11.10 84.06 ±15.65 |
GG AA GA |
1.059 |
|
Control
|
27.40 ± 4.27 38.00 ± 6.59 29.80 ± 3.08 |
GG AA GA |
2.76 |
|
ESRD: End Stage Renal Disease; SD: Standard Deviation; P ≤ 0.05. |
|||
Discussion
The importance of renalase in regulating blood pressure calls for further investigation into the causes and potential treatments for hypertension, which is increasingly common among hemodialysis patients [13]. There is an SNP called rs2576178 that is located in the 5'-flanking regions of the RNLS gene. It replaces a single nucleotide with another, potentially influencing the regulation and expression of the RNLS gene [12].
It is already known that changes in the renalase gene (rs2576178 SNP) are connected to high blood pressure, heart disease, stroke, ESRD, coronary cardiopathy, and naturally occurring high blood pressure [14]. Patients with end-stage renal illness had a frequency of 0.32 for the rs2576178 SNP minor, G allele, while the healthy control group had a frequency of 0.34. These investigations' findings have been inconclusive for the patient and control groups. The frequency of the minor G allele was 0.34 in the dbSNP database. In 2011, Gambaro et al. [15] discovered that 0.35 of Egyptians with a parathyroid gland had the minor G allele frequency for the rs2576178 SNP. This was a little different from what we found. Li et al. [16] discovered that systolic and diastolic blood pressure values differed significantly depending on genotype, and that individuals with parathyroid glands had a significantly higher GG genotype of the rs2576178 SNP [17,18].
When comparing ESRD patients to healthy controls, the present study did not find a statistically significant correlation (P 0.05) between blood renalase levels and rs2576178 genotype frequencies. This could mean that the GG genotype (rs2576178 SNP) in the 5'-flanking region of the RNLS gene, which was found to have a big effect on gene expression before, either doesn't have much of an effect on renalase expression or doesn't have any effect at all. Other single-nucleotide polymorphisms (SNPs) in the RNLS gene, namely rs10887800 and rs2296545, are usually linked to high blood pressure and may help explain why patients with kidney disease have higher serum renalase levels. While we did test serum renalase levels, we did not measure enzyme activity in our experiment. According to our results, high levels of renalase in the blood of people with renal failure—most of whom also had high blood pressure—may be because the enzyme cannot break down the catecholamines (epinephrine and noradrenaline) that it needs as substrates. This, in turn, raises blood pressure. Thus, it is necessary to assess the level and activity of serum renalase in patients with renal insufficiency and hypertension.
When the rs2576178 SNP in the 5'-flanking region of the RNLS gene was tested on people with end-stage renal failure who were on dialysis, there were no big differences at the GG genotype. Consistent with earlier studies, this study did not detect a difference in the frequency of the G allele or the GG genotype between HD patients and contributors [19]. A highly Swedish population review also failed to find a correlation between upset and the SNP rs2576178 [20]. Additionally, Ahlawat et al. [21] found no link between the RNLS gene (rs2576178) polymorphism and the bothersome, long-lasting renal failure illness, based on genotype delivery and gene frequencies at SNP rs2576178.
Lastly, renal disorders are complex diseases involving multiple factors. These factors include inflammation, nutrition, environmental factors, and a large number of genes that are involved in renal functions or have risk variants that could worsen the disease.
Conflicts of Interest
The author states that they have no conflicts of interest.
Data Availability
None.
Funding Source
None.
Ethics Approval
Ethical clearance was granted for the study by the Babylon Health Directorate in Babil, Iraq, with reference number 246. Research conducted in accordance with the Declaration of Helsinki standards.
Authors' Contribution
Zahraa Isam Jameel and her colleagues came up with the concept and wrote, reviewed, and edited the final version.
Acknowledgements
For providing the necessary materials, the writers would like to thank the College of Science at Babylon University.
References
2. Desir GV, Wang L, Peixoto AJ. Human renalase: a review of its biology, function, and implications for hypertension. J Am Soc Hypertens. 2012 Nov-Dec;6(6):417-26.
3. Desir GV. Renalase deficiency in chronic kidney disease, and its contribution to hypertension and cardiovascular disease. Curr Opin Nephrol Hypertens. 2008 Mar;17(2):181-5.
4. Hennebry SC, Eikelis N, Socratous F, Desir G, Lambert G, Schlaich M. Renalase, a novel soluble FAD-dependent protein, is synthesized in the brain and peripheral nerves. Mol Psychiatry. 2010 Mar;15(3):234-6.
5. Boomsma F, Tipton KF. Renalase, a catecholamine-metabolising enzyme? J Neural Transm (Vienna). 2007;114(6):775-6.
6. Desir GV. Regulation of blood pressure and cardiovascular function by renalase. Kidney Int. 2009 Aug;76(4):366-70.
7. Farzaneh-Far R, Desir GV, Na B, Schiller NB, Whooley MA. A functional polymorphism in renalase (Glu37Asp) is associated with cardiac hypertrophy, dysfunction, and ischemia: data from the heart and soul study. PLoS One. 2010 Oct 20;5(10):e13496.
8. Guo X, Wang L, Velazquez H, Safirstein R, Desir GV. Renalase: its role as a cytokine, and an update on its association with type 1 diabetes and ischemic stroke. Curr Opin Nephrol Hypertens. 2014 Sep;23(5):513-8.
9. Malyszko J, Bachorzewska-Gajewska H, Dobrzycki S. Renalase, kidney and cardiovascular disease: are they related or just coincidentally associated?. Advances in Medical Sciences. 2015 Mar 1;60(1):41-9.
10. Stec A. Rs10887800 renalase gene polymorphism influences the level of circulating renalase in patients undergoing hemodialysis but not in healthy controls. BMC Nephrology. 2017 Dec;18:1-6.
11. Zhang F, Liu W, Wu Y, Li X, Zhang S, Feng Y, et al. Association of renalase gene polymorphisms with the risk of hypertensive disorders of pregnancy in northeastern Han Chinese population. Gynecological Endocrinology. 2020 Nov 1;36(11):986-90.
12. Buraczynska M, Zukowski P, Buraczynska K, Mozul S, Ksiazek A. Renalase gene polymorphisms in patients with type 2 diabetes, hypertension and stroke. Neuromolecular Med. 2011 Dec;13(4):321-7.
13. Kiseljakovic E, Mackic-Djurovic M, Hasic S, Beciragic A, Valjevac A, Alic L, et al. Renalase Gene rs2576178 Polymorphism in Hemodialysis Patients: Study in Bosnia and Herzegovina. Med Arch. 2016 Feb;70(1):31-4.
14. Stec A, Semczuk A, Furmaga J, Ksiazek A, Buraczynska M. Polymorphism of the renalase gene in end-stage renal disease patients affected by hypertension. Nephrology Dialysis Transplantation. 2012 Nov 1;27(11):4162-6.
15. Gambaro G, Soldati L, Vezzoli G. Genetics and molecular biology of renal stones. In: Rao N, Preminger G, Kavanagh J, (eds). Urinary Tract Stone Disease. London: Springer; 2010 Dec 2, pp. 9-15.
16. Li G, Xu J, Wang P, Velazquez H, Li Y, Wu Y, et al. Catecholamines regulate the activity, secretion, and synthesis of renalase. Circulation. 2008 Mar 11;117(10):1277-82.
17. Isam Z, Omran R, Mahmood AH. Single-nucleotide polymorphisms of calcium-sensing receptor encoding gene associated with calcium kidney stone disease in Babylon province. Asian J Pharm Clin Res. 2018;11(2):417-21.
18. Isam ZA, Al-jelawi RO, Mahmood AH. Detection Single Nucleotide Polymorphisms in Uromodulin Promoter Region Associated with Renal Diseases Using Sin-gle-Strand Conformation Polymorphism-Polymerase Chain Polymorphisms Technique. Asian J Pharm Clin Res. 2003;11:205.
19. Abdallah ES, Sabry D. Renalase gene polymorphisms in end-stage renal disease patients: an Egyptian study. J Am Sci. 2013;9(1): 346-9.
20. Fava C, Montagnana M, Danese E, Sjögren M, Almgren P, Engström G, et al. The Renalase Asp37Glu polymorphism is not associated with hypertension and cardiovascular events in an urban-based prospective cohort: the Malmö Diet and cancer study. BMC Med Genet. 2012 Jul 19;13:57.
21. Ahlawat R, Gupta S, Kapoor S, Kar P. Polymorphism of Renalase Gene in Patients of Chronic Kidney Disease. Open Journal of Nephrology. 2012;2(4):136-43.