Abstract
The dimer form Pyruvate kinase M2 (PKM2) possesses lower glycolytic ability compared to its terameter form. This is of great importance for cancer cells since dimer PKM2 increases the levels of glycolytic intermediates for cell proliferation and growth. Accumulating evidence has also revealed that dimer PKM2 has many non-glycolytic functions including acting as a transcription factor and protein kinase. Our recent work has uncovered a novel non-glycolytic function of dimer PKM2. PKM2 coordinates the switch between glycolysis and glutaminolysis in cancer cells. We further discovered that dimer PKM2 upregulates glutaminolysis through internal ribosome entry site (IRES)-dependent translation of c-Myc. Here in this commentary, we discuss several most important unanswered questions relevant to this new finding, in hope to inspire future studies. We discussed several possibilities that dimer PKM2 regulates IRES-dependent translation of other genes as well as the role(s) of dimer PKM2 in regulation of glutaminolysis in non-cancer cells. Furthermore, we also discuss the potential mechanism of PKM2 in regulating IRES-dependent translation of c-Myc.
Keywords
Pyruvate kinase M2, IRES (Internal ribosome entry site), Glutaminolysis, c-Myc
Commentary
Cancer cells reprogram their nutrition metabolism to meet their high bioenergetic and biosynthetic demands in support of rapid growth and continuous proliferation [1]. Both glycolysis and glutaminolysis are altered in cancer cells [2]. Glycolysis is a universal pathway used by all living cells for energy production, which can occur in both aerobic and anaerobic states. However, about a century ago, Otto Warburg first discovered that the glycolysis rate is ~200 times higher in cancer cells compared to healthy cells and cancer cells predominantly undergo anaerobic glycolysis regardless of whether oxygen is present [2,3]. This phenomenon is later termed Warburg effect. After its discovery, scientists have come up with many theories for why cancer cells switch to inefficient anaerobic glycolysis. One widely accepted theory is that Warburg effect allows cancer cells to maintain large pools of intermediates which provide building blocks for synthesis of nucleotides, lipids and amino acid, assisting the rapid proliferation and growth of cancer cells [4].
Warburg effect limits entry of pyruvate into TCA cycle in cancer cells. To compensate for the reduction in carbon coming from glycolysis, cancer cells switch to the glutaminolysis which utilizes glutamine to replenish the TCA cycle [5]. In addition, glutaminolysis also provides reducing power by converting glutamate to glutathione (GSH), the most abundant anti-oxidant in mammalian cells in handling oxidative stress [5]. Elevated glutaminolysis has been recognized as a critical hallmark of cancer [6]. Not only that cancer patients have much lower concentrations of glutamine in their blood circulation compared to healthy individual [6], but also, the reduction in glutamine consumption can restrain the cancer progression to some extent [7]. Glutamine, the most abundant amino acid in blood circulation, is beneficial for cancer cells as it supplies more carbon and nitrogen for macromolecule biosynthesis and maintenance of redox balance [8-10]. By this means, with increased glutamine being converted via glutaminolysis, cancer cells display enhanced cell proliferation rate and reduced cell death [11].
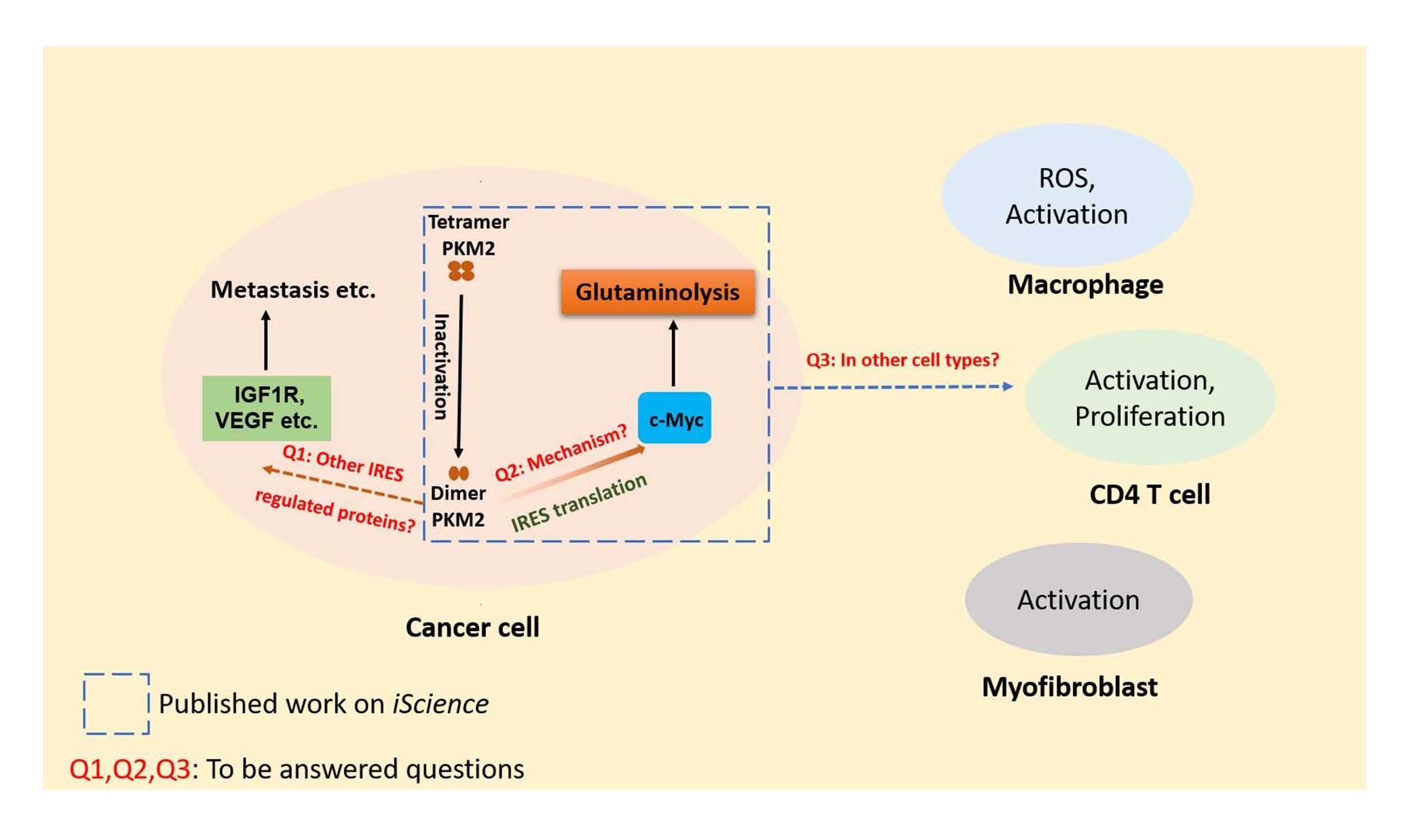
Glutaminolysis as well as glycolysis in cancer cells have been extensively studied, but relatively less attention has been paid to how the cancer cells coordinate these two metabolic pathways. In a recent manuscript entitled “Pyruvate kinase M2 coordinates metabolism switch between glycolysis and glutaminolysis in cancer cells” [12], the authors demonstrated that the pyruvate kinase M2 (PKM2), a critical enzyme in glycolysis, helps to switch the glutaminolysis pathway in cancer cells through internal ribosome entry site (IRES)-dependent translation of c-Myc. This study uncovers an important mechanism how cancer cells coordinate glycolysis and glutaminolysis and also sheds light on the novel role of PKM2 as a regulator of glutaminolysis. Nevertheless, the unique way of PKM2 in regulation of glutaminolysis in cancer cells also brings up some intriguing questions which are worthy of further investigations (Figure 1): Does PKM2 manipulate any other gene expression through IRES-dependent translation? Does PKM2 regulates glutaminolysis in other cell types besides cancer cells? What is the exact role of PKM2 in IRES-dependent translation of c-Myc?
Figure 1. Questions raised by new function of dimer PKM2 in regulation of glutaminolysis in cancer cells. Dimer PKM2 coordinates glutaminolysis through the regulation of c-Myc via IRES-dependent translation. Questions (Q1, Q2 and Q3) are raised by this newly discovered role of dimer PKM2. Q1: Does PKM2 manipulate other proteins through IRES-dependent translation? Q2: What is the exact role of PKM2 in the IRES-dependent translation of c-Myc? Q3: Does PKM2 regulate glutaminolysis in other cell types besides cancer cells?
The Unique Role of PKM2 in Regulation of Glutaminolysis
Pyruvate kinase is a key enzyme in glycolysis, which catalyzes the conversion of phosphoenolpyruvate (PEP) to pyruvate at the last, yet a rate-limiting, step of glycolysis. Although there are several different isoforms of pyruvate kinase, cancer cells highly express M2 isoform [13]. More interestingly, PKM2 in cancer cells is predominantly enzymatic inactive form, dimer PKM2 [14]. Compared to its active tetramer PKM2, the low-activity-dimer PKM2 has compromised affinity for its substrate PEP, leading to accumulation of glycolytic intermediates. Despite its glycolytic roles, PKM2 in cancer cells have additional non-glycolytic functions [14,15]. For instance, PKM2 in cancer cells is identified to regulate HIF1α activity as a transcriptional activator [16]; and PKM2 is also demonstrated to phosphorylate STAT3 using PEP as a phosphate donor in cancer cells [17]. These various noncanonical functions suggest that roles of PKM2 in cancer cells are complicated, and there might be other nonglycolytic functions which have not been fully elucidated. Here in the new study “Pyruvate kinase M2 coordinates metabolism switch between glycolysis and glutaminolysis in cancer cells”, the authors uncovered another novel nonglycolytic function of PKM2 in cancer cells.
In recent study, Li and co-workers demonstrated the mechanism how PKM2 coordinates the glycolysis and glutaminolysis pathways. The authors showed for the first time a crucial role of inactive dimer PKM2 in regulating IRES-dependent protein translation in cancer cells. This study demonstrated that PKM2 dimer, promoted by growth stimulations, elevates glutaminolysis by upregulating mitochondrial glutaminase I (GLS-1) via interacting with c-myc IRES-RNA and facilitating IRES-dependent translation of c-Myc. The traditional understanding of the role of PKM2 in cancer metabolism is that the inactive dimer PKM2 support cancer cell growth by allowing the glycolytic intermediates to build-up and re-direct into anabolic pathways [18], an advance made by the latest report is that dimer PKM2 can also modulate the augmentation of glutaminolysis, feeding bio-synthesis materials to cancer cells.
Could Dimeric PKM2 Regulate IRESdependent Translation of Other Proteins?
A number of proliferation and survival related genes are regulated at translational level [19]. This is important to cancer cells since translational regulation provides them rapid adjustment to stressful environment [20]. Accumulating evidence shows that IRES-dependent translation is important for many cancer-related proteins [21-23], suggesting that IRES-mediated translation is vital for cancer cells. Our work showed that PKM2 can regulate GLS-1 through IRES-dependent translation of c-Myc. Since c-Myc is an important regulator in cancer cells [24], PKM2 might regulate other c-Myc related pathways such as cell cycle, apoptosis and proliferation [25-28], which are worth further investigations.
In addition to c-Myc, many other cancer-progression associated genes have been reported to be regulated by IRES-mediated translation, such as VEGF, IGF1R, cyclin D1 and XIAP [29-33], Furthermore, IRES-mediated translation of these genes have been found critical for many processes of cancer cells including metastasis and angiogenesis [31,34,35]. Interestingly, dimer PKM2 is found to promote cancer cell metastasis and angiogenesis [36,37]. It is plausible that the dimer PKM2 might be able to regulate these genes through IRES-dependent translation. It is worthy to further test if PKM2 plays a role in regulation of these cancer promoting genes through IRES-dependent translation in cancer cells, which may suggest a broader implication of dimer PKM2 in reprogramming and coordinating multiple cellular processes in cancer cells.
Could Dimeric PKM2 Regulate Glutaminolysis in Non-cancer Cell Types?
Besides tumor cells, metabolic reprogramming of glutaminolysis is observed in multiple cell types such as immune cells [38-40]. The role of dimer PKM2 in regulation of glutaminolysis in non-cancer cell types merits further investigations.
Macrophages and neutrophils stimulated by LPS or bacterial infection utilize glutaminolysis to sustain ROS production by generating NADH [41]. Furthermore, macrophages and neutrophils upon microbial activation display an increased level of PKM2 expression [42,43] and PKM2 in LPS-activated macrophages are primarily dimers [43]. Altogether, innate immune cells such as macrophages following microbial challenge might elevate dimer PKM2 to rewire glutaminolysis through a similar mechanism as was observed in cancer cells. Noticeably, the role of dimer PKM2 in macrophages might not act through the regulation of IRES-dependent translation of c-Myc as c-Myc can drive macrophages to alternative activated phenotype [44] which mainly have tetramer PKM2 [43]. Nevertheless, it is still worthy to investigate whether dimer PKM2 can regulate the glutaminolysis in innate immune cells and the underneath mechanisms if it can.
Besides the innate immune cells, it is well appreciated that dimer PKM2 is essential for CD4+ T cell activation [45]. Moreover, CD4+ T cell activation also relies on glutaminolysis and c-Myc is an indispensable factor in this process [46-48]. So, it is possible that dimer PKM2 mediates glutaminolysis activation in the process of T cell activation via regulating c-Myc by IRES-dependent translation.
In addition to immune cells, emerging evidence suggests that glutaminolysis is required for the activation of myofibroblasts [49,50], whose persistence is a hallmark of fibrotic diseases of multiply tissues and organs [50]. PKM2 is expressed in fibroblasts. Thus, it is interesting and important to analyze potential role of dimer PKM2 in gluaminolysis regulation in fibroblasts differentiation and activation to myofibroblasts, which may shed light on the molecular mechanism of organ and tissue fibrosis development and progression.
What are the Possible Roles of PKM2 in IRES-dependent Translation of C-Myc?
Dimer PKM2 regulates c-Myc expression via the IRESdependent translation. However, the detailed mechanism about how PKM2 regulates c-Myc expression via IRESdependent translation remains unclear. IRES-dependent translation involves multiple different proteins [22,51]. Inferring from the observations, it is plausible that PKM2 may facilitate assembly of hnRNP L/K into c-myc IRES complex. However, further study is needed to elucidate how PKM2 regulates the assembly process of c-myc IRES complex. In addition, it is also unknown whether dimer PKM2 is directly involved in the IRES-dependent c-Myc translation. Since the recruitment of translation initiation factors to IRES sequence and structure is critical for IRES-dependent translation [52,53]. It is reasonable to hypothesize that dimer PKM2 may be directly involved in recruiting IRES associated translation initiation factors and/or modulating the IRES structure, which could be deciphered with further investigations.
References
2. Akins NS, Nielson TC, Le HV. Inhibition of glycolysis and glutaminolysis: an emerging drug discovery approach to combat cancer. Current Topics in Medicinal Chemistry. 2018 Mar 1;18(6):494-504.
3. Warburg O, Posener K, Negelein E. Über den stoffwechsel der carcinomzelle. Naturwissenschaften. 1924 Dec 1;12(50):1131-7.
4. Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends in Biochemical Sciences. 2016 Mar 1;41(3):211-8.
5. Anderson NM, Mucka P, Kern JG, Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein & Cell. 2018 Feb;9(2):216-37.
6. Cluntun AA, Lukey MJ, Cerione RA, Locasale JW. Glutamine metabolism in cancer: understanding the heterogeneity. Trends in Cancer. 2017 Mar 1;3(3):169-80.
7. Jiang J, Srivastava S, Zhang J. Starve cancer cells of glutamine: break the spell or make a hungry monster? Cancers. 2019 Jun;11(6):804.
8. Souba WW. Glutamine and cancer. Annals of Surgery. 1993 Dec;218(6):715-28.
9. Kodama M, Oshikawa K, Shimizu H, Yoshioka S, Takahashi M, Izumi Y, et al. A shift in glutamine nitrogen metabolism contributes to the malignant progression of cancer. Nature Communications. 2020 Mar 17;11(1):1320.
10. Alberghina L, Gaglio D. Redox control of glutamine utilization in cancer. Cell Death & Disease. 2014 Dec;5(12):e1561.
11. Matés JM, Pérez-Gómez C, de Castro IN, Asenjo M, Márquez J. Glutamine and its relationship with intracellular redox status, oxidative stress and cell proliferation/death. The International Journal of Biochemistry & Cell Biology. 2002 May 1;34(5):439-58.
12. Li L, Peng G, Liu X, Zhang Y, Han H, Liu ZR. Pyruvate Kinase M2 Coordinates Metabolism Switch between Glycolysis and Glutaminolysis in Cancer Cells. Iscience. 2020 Nov 20;23(11):101684.
13. Prakasam G, Iqbal MA, Bamezai RN, Mazurek S. Posttranslational modifications of pyruvate kinase M2: tweaks that benefit cancer. Frontiers in Oncology. 2018 Feb 7;8:22.
14. Zahra K, Dey T, Ashish A, Pandey U, Mishra SP. Pyruvate kinase M2 and cancer: the role of PKM2 in promoting tumorigenesis. Frontiers in Oncology. 2020;10:159.
15. Hsu MC, Hung WC. Pyruvate kinase M2 fuels multiple aspects of cancer cells: from cellular metabolism, transcriptional regulation to extracellular signaling. Molecular Cancer. 2018 Dec;17(1): p. 35.
16. Azoitei N, Becher A, Steinestel K, Rouhi A, Diepold K, Genze F, et al. PKM2 promotes tumor angiogenesis by regulating HIF-1α through NF-κB activation. Molecular Cancer. 2016 Dec;15(1): p.3.
17. Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Molecular Cell. 2012 Mar 9;45(5):598-609.
18. Dang CV. Links between metabolism and cancer. Genes & Development. 2012 May 1;26(9):877-90.
19. Ruggero D. Translational control in cancer etiology. Cold Spring Harbor Perspectives in Biology. 2013 Feb 1;5(2):a012336.
20. Liu B, Qian SB. Translational reprogramming in cellular stress response. Wiley Interdisciplinary Reviews: RNA. 2014 May;5(3):301-5.
21. Xi S, Zhao M, Wang S, Ma L, Wang S, Cong X, et al. IRES-mediated protein translation overcomes suppression by the p14ARF tumor suppressor protein. Journal of Cancer. 2017;8(6):1082-88.
22. Yang Y, Wang Z. IRES-mediated cap-independent translation, a path leading to hidden proteome. Journal of Molecular Cell Biology. 2019 Oct;11(10):911-9.
23. Grzmil M, Hemmings BA. Translation regulation as a therapeutic target in cancer. Cancer Research. 2012 Aug 15;72(16):3891.
24. Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clinical cancer research: an official journal of the American Association for Cancer Research, 2012. 18(20): p. 5546-5553.
25. McMahon SB. MYC and the control of apoptosis. Cold Spring Harbor perspectives in medicine. 2014 Jul 1;4(7):a014407.
26. Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Molecular and Cellular Biology. 1999 Jan 1;19(1):1-11.
27. Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008 Oct;27(50):6462-72.
28. Desbarats L, Schneider A, Müller D, Bürgin A, Eilers M. Myc: a single gene controls both proliferation and apoptosis in mammalian cells. Experientia. 1996 Dec;52(12):1123-9.
29. Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell cycle. 2011 Jan 15;10(2):229-40.
30. Godet AC, David F, Hantelys F, Tatin F, Lacazette E, Garmy-Susini B, et al. IRES trans-acting factors, key actors of the stress response. International Journal of Molecular Sciences. 2019 Jan;20(4):924.
31. Kucab JE, Dunn SE. Role of IGF-1R in mediating breast cancer invasion and metastasis. Breast Disease. 2003 Jan 1;17(1):41-7.
32. Loges S, Mazzone M, Hohensinner P, Carmeliet P. Silencing or fueling metastasis with VEGF inhibitors: antiangiogenesis revisited. Cancer Cell. 2009 Mar 3;15(3):167-70.
33. Vaklavas C, Meng Z, Choi H, Grizzle WE, Zinn KR, Blume SW. Small molecule inhibitors of IRES-mediated translation. Cancer Biology & Therapy. 2015 Oct 1;16(10):1471-85.
34. Walters B, Thompson SR. Cap-independent translational control of carcinogenesis. Frontiers in Oncology. 2016 May 25;6:128.
35. Vaklavas C, Blume SW, Grizzle WE. Translational dysregulation in cancer: molecular insights and potential clinical applications in biomarker development. Frontiers in Oncology. 2017 Jul 26;7:158.
36. He X, Du S, Lei T, Li X, Liu Y, Wang H, et al. PKM2 in carcinogenesis and oncotherapy. Oncotarget. 2017 Dec 15;8(66):110656-70.
37. Li L, Peng G, Liu X, Zhang Y, Han H, Liu ZR. Pyruvate Kinase M2 Coordinates Metabolism Switch between Glycolysis and Glutaminolysis in Cancer Cells. Iscience. 2020 Nov 20;23(11):101684.
38. Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013 Apr 18;38(4):633-43.
39. Kim J. Regulation of immune cell functions by metabolic reprogramming. Journal of Immunology Research. 2018 Feb 13;2018; p. 8605471-8605471.
40. Wu S, Kuang H, Ke J, Pi M, Yang DH. Metabolic Reprogramming Induces Immune Cell Dysfunction in the Tumor Microenvironment of Multiple Myeloma. Frontiers in Oncology. 2021 Jan 15;10:3061.
41. Zhang X, Zink F, Hezel F, Vogt J, Wachter U, Wepler M, et al. Metabolic substrate utilization in stress-induced immune cells. Intensive Care Medicine Experimental. 2020 Dec;8(1):p. 28.
42. Zhang Y, Li L, Liu Y, Liu ZR. PKM2 released by neutrophils at wound site facilitates early wound healing by promoting angiogenesis. Wound Repair and Regeneration. 2016 Mar;24(2):328-36.
43. Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metabolism. 2015 Jan 6;21(1):65-80.
44. Pello OM, De Pizzol M, Mirolo M, Soucek L, Zammataro L, Amabile A, et al. Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood. 2012 Jan 12;119(2):411-21.
45. Angiari S, Runtsch MC, Sutton CE, Palsson-McDermott EM, Kelly B, Rana N, et al. Pharmacological activation of pyruvate kinase M2 inhibits CD4+ T cell pathogenicity and suppresses autoimmunity. Cell Metabolism. 2020 Feb 4;31(2):391-405.e8.
46. Araujo L, Khim P, Mkhikian H, Mortales CL, Demetriou M. Glycolysis and glutaminolysis cooperatively control T cell function by limiting metabolite supply to N-glycosylation. Elife. 2017 Jan 6;6:e21330.
47. Wahl DR, Byersdorfer CA, Ferrara JL, Opipari Jr AW, Glick GD. Distinct metabolic programs in activated T cells: opportunities for selective immunomodulation. Immunological Reviews. 2012 Sep;249(1):104-15.
48. Bandukwala HS, Gagnon J, Togher S, Greenbaum JA, Lamperti ED, Parr NJ, et al. Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proceedings of the National Academy of Sciences. 2012 Sep 4;109(36):14532-7.
49. Gibb AA, Lombardi AA, Murray EK, Tan Y, Lorkiewicz PK, Huynh AT, et al. Glutaminolysis is Required to Initiate Myofibroblast Differentiation and Persistence During Stress. Circulation Research. 2019 Aug 2;125(Suppl_1):A517.
50. Bernard K, Logsdon NJ, Benavides GA, Sanders Y, Zhang J, Darley-Usmar VM, et al. Glutaminolysis is required for transforming growth factor-β1–induced myofibroblast differentiation and activation. Journal of Biological Chemistry. 2018 Jan 26;293(4):1218-28.
51. Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Development. 2001 Jul 1;15(13):1593-612.
52. Fitzgerald KD, Semler BL. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2009 Sep 1;1789(9-10):518-28.