Abstract
Breast cancer is one of the most diagnosed cancers around the world. Metastases are the leading cause of death related to breast cancer. It has been noticed, in recent years, that non-coding long RNAs (lncRNAs) are strongly related to the development and progress of breast cancer. Current studies have noted that lncRNAs can be divided into two subgroups: those who are ongogenic and those who serve rather as tumor suppressors. This review analyzes the correlation of RNA with the development of breast cancer.
Keywords
Breast cancer, Oncogenes, lncRNAs
Introduction
At present, breast cancer is more frequently diagnosed in women than in men. According to global cancer statistics, each year more than 1,675,000 women are diagnosed and more than 500,000 of them die [1]. Some subtypes of breast cancer have been described. [5]
Breast cancer tumors have been classified into three main groups. Such classification is based on the gene expression of estrogen receptors (ER) and human epidermal growth factors (HER2). The most frequently diagnosed subtype is ER (positive tumors), followed by HER (positive tumors) (which houses ERBB2 amplification) and ER, HER2 and triple-negative progesterone receptors in tumors [2].
With regard to diagnosis and therapies against breast cancer, it is necessary to work on new approaches for improvement, understanding of molecular mechanisms is the most important the for this purpose. It has been found in in vitro experiments that non-coding long RNAs may have key and important tasks in the development of breast cancer tumors [4,5], in addition to idealizing innovative proposals for new therapies, such as biomarkers.
General Characteristics of LncRNAs
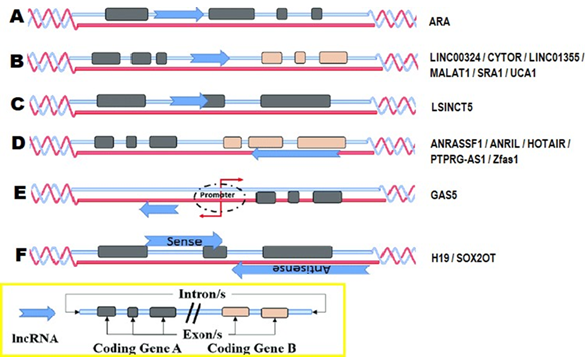
LncRNAs can be defined as RNA transcripts above 200 bp, which have no open reading frames. LncRNAs have complicated structures and intrinsic origins. It is not possible to define them by their length or inability to encode proteins. It has been observed that lncRNAs in chromatin state have preserved domains, be those marked by H3k4me3 and H3k36me3 [6]. LncRNAs can be classified according to their origin in the genome, as seen in Figure 1. Five groups are distinguished: Sense lncRNA, are those where the sequence of the lncRNAs overlap with one or more exons of another reading frame in the same direction. Then we have the Antisense lncRNA, in which it happens analogously to the Sense lncRNA, but this time in the opposite direction. The next category is that of bidirectional lncRNA, where the lncRNA begins in close genomic proximity with a neighboring reading frame, in the opposite chain. The intrinsic lncRNA are derived completely from an intron of another reading frame. Sometimes they can represent the sequence before the messenger. Finally, intergenic lncRNA, which derive from a sequence within a genomic interval, between two genes. [7]
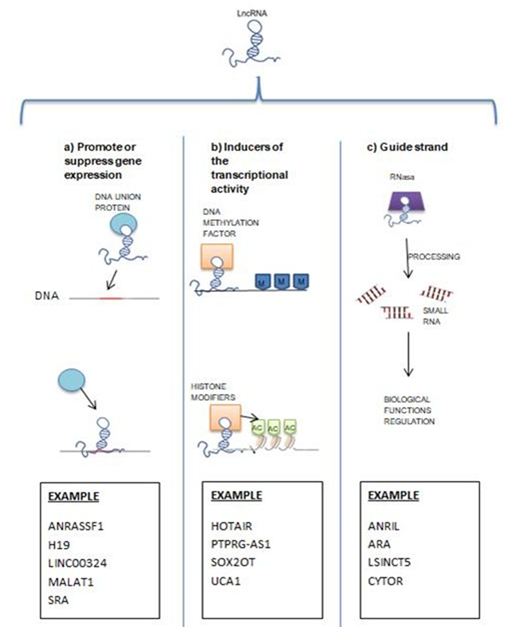
The lncRNAs are related to several biological and physiological processes, with different functions for each case. They are shown to be important regulators as tumor suppressors, as well as suppressors of oncogenesis pathways. Current studies argue that lncRNA are responsible for regulating the major pathways leading to cancer at three different levels: epigenetic, transcriptional, and post-transcriptional [6]
The best known and potential function of RNAin is related to the epigenetic regulation of white genes.
Primarily because of its repressive functions. A wide range of lncRNAs such as ANRIL, HOTAIR, H19 and XIST have been shown to rebuke transcription by histone modification or chromatin protein remodeling [8]. Noncoding RNAs can act on cis and trans systems. For cis, these are restricted to the site of synthesis or chromosome and tend to regulate only a few genes, while trans-acting RNAs spread to other chromosomes and regulate a wider range of genes [9].
Mechanism of lncRNAs in Cancer Biology
Notably deregulated lncRNA has been found in tumor tissues, in previous studies their participation in the development and progression of various types of cancer is established [10,11]. There are lncRNAs that participate in 3 different levels in biological processes: transcriptional, post-transcriptional and epigenetic level (Figure 1), these lncRNAs lack open reading frame (ORF).
Transcriptional level of regulation by lncRNAs
lncRNA are transcribed from active enhancers: The presence of nearby promoters is necessary for the transcription of most genes, these promoters have more distant enhancer elements. Enhancers are located distant from the transcription initiation site (TSS) most of the time and bind to tissue-specific TF, thus functioning as regulators of differential gene expression [12]. The combined effects of one or more enhancers almost always regulate the expression of a particular gene, these enhancers are activated at a specific site, key to both general and tissue development. RNAPII allows binding between enhancers, reflecting their interaction with the promoter. Neuron studies of mice by Kim et al [13] on the activation of calcium signaling, found that an approximately 2 kb ncRNA is transcribed from bidirectionally active enhancers.
LncRNA genes that have an enhancer activity:
NcRNAs are transcribed from independent locus, but not from enhancers, they are a type of lncRNA with transcriptional activation functions [14-16]. It is responsible for the activation of specific transcription of neighboring genes in a particular RNA, requiring the activity of a promoter [14]. Through a chromosomal conformational change, the gene activation is mediated by an ncRNA, allowing an approach between the ncRNA locus and the promoter of its target gene [17]. A protein complex, known as a mediator, together with cohesin, is part of the binding of promoters and enhancers [18].
Post transcriptional regulation by lncRNAs
LncRNA as a source of miRNA: From a primary transcript through a two-stage process, miRNA (primiRNA) is originated. The pri-miRNA is cut while it is still in the nucleus through chromosomal region 8 in Drosha and Digeorge syndrome (DGCR8). The premiRNA is transferred to the cytoplasm as a consequence of the cut, in the cytoplasm it is processed thanks to the Dicer / TAR RNA protein (TRBP), finally producing a mature miRNA [19], [20]. Almost all pri-miRNA have a length of more than 1 kb [20], and can be considered lncRNA. There are two main sources of pri-miRNA in the genome: those whose expression is linked to the expression of an original transcript, although it does not always occur, caused by incorporation into another gene. There are also those that are transcribed independently of what was previously considered an intergenic region. The other group contains self-regulating promoters through RNAPII, similar to mRNA [21].
Approximately 50% of miRNAs are obtained from noncoding transcripts [22]. Many miRNAs are present in non-coding genes and are also located in introns [21]. From this genomic organization, it is proposed that the host lncRNA does not simply act as a pri-miRNA, but also have complementary roles encoded by exons, which is known as intronic concealment of lncRNAs to miRNAs. Some examples include DLEU2, which is the host gene of the miR-15a / 16.1 tumor suppressor group, located in its third intron [23,24]. Commonly miRNAs are shown infrared in leukemia. In adult chronic lymphocytic leukemia, it appears to be regulated by the host gene promoter that binds through transcription factors (TF), MYC [24] and PAX5 (formerly known as B-cell specific activator protein, BSAP) [25], since the expression of miR- 15a / 16.1, while acute myeloid leukemia in infants, the information obtained by methylation tests, indicates that this group of miRNA is regulated independently of its host gene [26]. A second example is the miR-31 tumor suppressor, which is not regulated in breast cancer. The miR-31 gene is incorporated into an intron of the LOC554202 lncRNA, and its transcription regulates through a methylated state of the host gene promoter [27].
LncRNA as a negative regulator of miRNA: miRNA are negative regulators of cell expression. The transcripts are directed through the union of a short initial sequence of 7 nt in the miRNA to a miRNA response element (MRE), which are also short. The link should not necessarily be complementary [28], complicating the prediction of miRNA objectives. Through computer predictions, it is suggested that a single miRNA targets hundreds of transcripts [28], although physiologically speaking, the number of target genes that are targets of a miRNA is almost much lower always [29]. This reflects the lack of success and the disconnect between the number of predicted targets and the number of current targets, causing it to be surprising that many of the lncRNAs contain predicted miRNA binding sites. This raises interesting theories, and that the function of a number of lncRNAs could through a sequestration of miRNAs, possibly, regulate the genetic expression, leading to a limitation in the concentration in the cell, thus causing the reduction of the set of miRNA available in the cell. Which suggests that an lncRNA may act as a negative regulator for miRNA function and may be a possible positive regulator of genetic expression. This is known as the “competitive endogenous RNA” (ceRNA) hypothesis [30].
Epigenetic regulation, lncRNAs and cancer
Cancer in humans forms a lot of diseases that involve various genetic and epigenetic alterations. The change from silenced genes to active and transcribable genes, and vice versa, is regulated by cumbersome mechanisms and complex alterations in cellular machinery [30]. Although only part of DNA mutations belongs to malignant phenotypes, recent studies have demonstrated the relevance of epigenetic alterations in tumor development [31]. Cancer cells show changes in DNA methylation patterns, such as global hypomethylation and CpG islands, in promoter regions. Due to these variations, the genome of cancer cells presents chromosomal fluctuation, loss of genomic imprinting and changes in gene expression, both for protein coding and for regulatory ncRNAs [32].
Thanks to the advance of high-performance sequencing, a variety of studies have provided a constantly expanding survey of genetic aberrations in cancer. Although, these deformations also affect the lncRNAs, causing the repression of their functions and, consequently, leading to a deregulation of their objectives [32]. Most of the well-characterized lncRNAs to date, show a functional role in the regulation of gene expression, in particular, transcriptionally rather than post-transcriptional. With the progress in the cancer transcriptome profile and the evidence gathered on the functions of lncRNA, a variety of lncRNA with differential expression have been related to different types of cancer, in addition, one or more dynamic changes in their structures are involved. [29]
LncRNAs in Breast Cancer
It has been found that different types of LncRNA are expressed in breast cancer; these are expressed abnormally in tumors [33,34]. LncRNAs are associated with breast cancer growth, because they are involved in the regulation of both cell proliferation and apoptosis. Some of the LncRNAs play an important role in the invasion, metastasis, and drug resistance of breast cancer [10]. In breast cancer several LncRNAs are involved [35]. Some of them have been studied and classified into two types: oncogenic type and tumor suppressor.
Oncogenic lncRNAs in breast cancer
There have been several oncological studies for the identification of LncRNA as functional genes associated with tumorigenesis and cancer development, these studies have revealed that some of the LncRNA have an oncogenic function because they are associated with distinctive characteristics of cancer, such as progression of the cell cycle, anti-apoptosis, angiogenesis and metastasis [36-37]. Some of the LncRNAs related to breast cancer are shown in Table 1 and are described below:
| Lnc RNA | Type of LncRNA | Function | Reference |
|---|---|---|---|
| ANRASSF1 | Antisense RNA | Promote proliferation | [38] |
| ANRIL | Antisense non coding RNA |
Promote proliferation | [39] |
| HOX | Antisense intergenic RNA | Metastasis | [45] |
| PTPRG- AS1 |
Overlapping, antisense | Differentially expressed between ER+ & ER- subtypes |
[28] |
| ARA | Intronic | Modulate MAPK signaling pathway, metabolism pathways, cell cycle and cell adhesion- related pathways. |
[29] |
| H19 | Overlapping | Induces growth arrest and apoptosis |
[46-48] |
| SOX2OT | Intronic lncRNA | Promote proliferation | [21-22] |
| LINC00324 | Long intergenic non- coding |
Canceringn Cell Marker | [28] |
| MALAT1 | Intergenic lncRNA | Metastasis | [30-32] |
| SRA | Intergenic lncRNA | Expression activator | [54-58] |
| LSINCT5 | Antisense lncRNA | Promote proliferation | [59] |
| UCA1 | Intergenic lncRNA | Metastasis | [61] |
| CYTOR | Intergenic lncRNA | Tumor marker | [24] |
ANRASSF1 (RNA 1 antisense RASSF1): It is an endogenous non-spliced RNA that is transcribed from the opposite chain in the tumor suppressor gene RASSF1. After studying, it has been revealed that it is overexpressed in the cell lines of breast and prostate tumors, unlike non-tumor cells since their expression is lower, but the presence of RASSF1A has an opposite pattern. It has been reported that the increase in ectopic expression of this LncRNA in HeLa cells causes a significant decrease in the expression of RASSF1A also shows an increase in cell proliferation [38].
ANRIL (Antisense non-coding RNA in the INK4 locus): Presents a great relation in the processes of tumorigenesis of several types of cancer, this has been demonstrated, thanks to the studies that have revealed its functioning through the regulation of its adjacent tumor suppressors CDKN2A/B through epigenetic mechanisms [39]. It also has several variants of splice, mostly these are polyadenylated. It has a great tissue specificity that is part of the splice variants and this is of great physiological importance in some tissues [40]. Studies showed that ANRIL is related to the response to ATM-dependent DNA damage, because it is elevated in DNA damage by transcription factor E2F1 [41].
The antisense intergovernmental RNA HOX (HOTAIR): It has an elevated expression in various malignant tumors, such as breast cancer [42]. In breast cancer HOTAIR competitively binds to the tumor suppressor BRCA1 [43-44], this union causes a transformation in epithelial cells. HOTAIR is overexpressed when there is an occurrence of primary and metastatic breast cancer compared to healthy breast epithelium, HOTAIR expression is implicated in the misdiagnosis of breast cancer and distant metastases [45].
PTPRG-AS1 (tyrosine phosphatase protein, receptor type, G, antisense): It is an antisense LncRNA which is complementary to the PTPRG tumor suppressor gene that is present in breast cancer. According to studies, PTPRG is part of the tyrosine phosphatase protein family that is linked to the regulation of cell growth and the mitotic cycle. Studies have shown that lower expression of PTPRG-AS1 is directly related to longer life expectancy in patients with breast cancer [28].
ARA (Adriamycin resistance associated): It is an intronic lncRNA, since it originates from an intron of the PAK3 gene. The relationship it presents with breast cancer is related to the sensitivity it has to adriamycin and overexpression in adriamycin-treated cell lines. It has been studied by transcriptomic analysis of microarrays, eliminating ARA causes a reduction in proliferation, an increase in cell death, G2 / M arrest and migration defects. ARA has a great modulating capacity in signaling pathways such as mitogen activated protein kinase (MAPK), metabolism pathways, cell cycle and pathways related to cell adhesion [29].
H19: It is one of the best studied LncRNA. It has a 2.3Kb transcript encoded by the maternal allele and it is capable of silencing the IGF2 gene in the same allele [19]. It is known that H19 is related to the control of embryonic growth and regulation of genetic impression [46,47]. IGF2 is under the control of H19 during embryogenesis [9]. MBD1 is a gene that presents an H3K9me3 modification of histone, this gene is recruited by H19 suppressing the genetic expression of some genes [48]. H19 has an elevated and abnormal expression in tumorigenic cells [49], due to this, it has been considered as an oncogenic RNA in breast epithelial cells [50]. In breast cancer cells, it was discovered that H19 was regulated by an increase in the S phase and its promoter was activated by E2F1 [51]. H19 is regulated by a widely deregulated transcription factor in cancer of epithelial origin (c-Myc) which is present in breast cancer. C- Myc binds to box E conserved in the H19 promoter and facilitates the initiation of transcription of H19 [52]. H19 has an important role in the phenotype of breast cancer tumors.
SOX2OT: The coding sequence of the SOX2 gene is found within the intron of the superimposed transcript of SOX2 (SOX2OT), which is a long multi-exon RNA [20]. In breast cancer, the expression of the SOX2 and SOX2OT gene is present, SOX2OT is performed with ER positive and negative ER breast cancer [21]. Both the gene and LncRNA are present in cancer stem cells and SOX2 was also studied which is a gene that participates in the maintenance of pluripotency [22]. Ectopic expression of SOX2OT increases the expression of the gene which in turn generates an increase in growth independent of breast cancer anchor [21].
LINC00324 (00324 non-long intergenic coding): LINC00324 is downstream of the CTC1 gene. It is known that CTC1 is involved in DNA replication and in the protection against telomere degradation. In LINC00324 breast cancer, it is expressed in the tumors and in ER type breast cancer. Overexpression of this gene is involved with the survival of patients with breast cancer [28].
MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript 1): MALAT1 is involved in the modulation of the alternative splice of pre-mRNAs [30]. In lung cancer, this LncRNA is connected with tumorigenesis and metastasis [53]. It has been found that there is an overexpression of MALTA1 in primary breast cancer tumors, in addition to that, MALTA1 has mutations and deletions in luminal breast cancer. These mutations have been found in the region that mediates the interaction with a splicing factor called SRSF1 [31].
SRA: The steroid receptor RNA activator (SRA) is known to be a lncRNA that has a function independent of the epigenetic or catalytic mechanism [54]. The SRA is involved with some hormonal receptors and this regulates the transaction of steroid receptor-dependent genes [56]. Overexpression of this LncRNA has been found in breast cancer [57], so it is considered an oncogene because its overexpression promotes cell proliferation [58].
LSINCT5: It is a stress-induced antisense lncRNA of 2.6KB. LSINCT5 is found in the nucleus and is transcribed by RNA polymerase III [59,60]; greater expression of LSINCT5 has been found in breast cancer and tumor tissues [59]; It has been found that reduced expression of LSINCT5 reduces cell proliferation in breast cancer tumors.
UCA1: Urothelial carcinoma-associated 1 (UCA1) is a 1.4Kb lncRNA. It is related to cell growth in breast cancer by suppressing p27, which is a tumor suppressor that inhibits cyclin- dependent kinase. The hnRNPI binding protein improves the translation of p27. UCA1 binds to this protein and prevents p27 from binding, so U27 suppresses p27. Likewise, this lncRNA has an oncogenic role in the proliferation of breast cancer [61].
CYTOR: It is an intergenic lncRNA located more than 100 kb away from the closest protein coding (PLGLB2) on chromosome 2p11.2. It has been discovered that it is regulated in several types of breast cancer, so it could be a tumor marker. It is involved in cell proliferation, cell migration, EMT and the EGFR, mTOR and MAPK pathways. CYTOR is associated with acetylation of H3K27, trimethylation of H3K4 and weak monomethylation marks of H3K4 in breast cancer [24].
lncRNA tumor auppressors in breast cancer
Some lncRNAs have been cataloged as tumor suppressors, research is still being done on this, so there are currently very few identified lncRNAs, among them few examples were shown in Table 2. These lncRNAs are involved in the DNA repair, as they have the ability to repair DNA damage. The following describes some tumor suppressor lncRNAs present in breast cancer:
| Lnc RNA | Type of LncRNA | Function | Reference |
|---|---|---|---|
| LINC01355 | Long intergenic non- coding RNA |
Suppresses the cell proliferation | [62] |
| GAS5 | Overlapping | It Is downregulation decreases breast cancer cell clonogenicity & anchorage- independent growth. |
[26] |
| Zfas1 | Overlapping, antisense |
Knockdown of Zfas1 in breast epithelial cells resulted in increased cell proliferation |
[64] |
| NBAT1 | Long intergenic non- coding RNA | Regulates cell proliferation and invasion by interacting with PRC2 (polycomb repressive complex 2) member EZH2 (enhancer of zeste 2) |
[65] |
| MEG3 | Downregulation | Repress PI3K/AKT/mTOR pathway and thus repressing the oncogenic behavior of the overly expressed miR-21 in BC cells |
[13] |
LINC01355: Under normal conditions, the expression of LINC01355 in breast cell lines is higher, in breast cancer, this lncRNA is underestimated. A low regulation of LINC01355 is associated with the size of the tumor and the TNM stage in breast cancer. The elimination of LINC01355 is related to the proliferation of breast cancer and the ectopic expression of LINC01355, this suppresses the proliferation and colony formation of breast cancer cells. When there is an overexpression of this apoptosis of lncRNA, it will increase in MCF7 xenograft tumors, because of this it is considered as a tumor suppressor in breast cancer [62].
Growth arrest-specific 5 (c): GAS5 is located at 1q25.1. A low regulation of GAS5 has been found in solid malignant neoplasms such as pancreatic cancers [26], colorectal [27], lung and breast. Studies on this lncRNA have revealed that it has a function as a tumor suppressor, this function is due to the fact that it sequesters several oncogenic miRNAs such as miR-221/222 [26], miR- 196. The low expression of GAS5 is involved both in the resistance to the drug tamoxifen [63] and in the resistance to dendrosomal curcumin in breast cancer cells. Among the most important research findings is that GAS5 is a regulator in the cell cycle that accumulates cells when they are in a state of growth arrest [26].
Zfas1: It is the antisense for the 5 ‘end of the protein coding gene, Znfx1, and the host snoRNA [64]. Zfas1 is found within the ducts and alveoli of the mammary gland, it was found that it is expressed differentially in the stage of pregnancy and lactation, due to this it is believed that it is involved in the development of the mammary gland. A low expression of Zfas1 in the epithelial cells of the breast generated an increase in cell proliferation. It has been found that in breast cancer the expression of this lncRNA is low [64].
Neuroblastoma associated transcript 1 (NBAT1): NBAT-1 is located at 6p22.3. A low regulation of NBAT1 has been found in different types of cancer, that’s the reason why it is known as a tumor suppressor lncRNA. NBAT1 is present in the regulation of cell proliferation and invasion since it interacts with the PRC2 member (complex repressive polyombomb 2) EZH2 (zeste 2 enhancer) [65]. A low expression of this lncRNA is involved with the tumor metastasis of the patient. Ectopic in vitro NBAT1 inhibits the migration and invasion of breast cancer cells. Some research has shown that NBAT1 is associated with the PRC2 member EZH2 and regulates the general gene expression profile; It is also known that DKK1 (WNT dikkop 1 signaling pathway inhibitor) is regulated by NBAT1 and its relationship with PRC2. NBAT1 is also involved in the inhibition of migration and invasion of breast cancer cells and, therefore, is considered as a possible inhibitor of metastasis in breast cancer [65].
Maternal expression 3 (MEG3): MEG3 is found chromosomally in 14q32.2. Low expression of MEG3 has been found in several malignant tumors. The survival of different patients with breast cancer has been involved with MEG3, so it is considered as a potential marker for breast cancer [66]. This lncRNA has an important role as a tumor suppressor because it has been found to act as a sponge for several miRNAs such as miR-9 [67], miR- 494 [68], miR-21 and miR-29 [12]. In breast cancer, its suppressive function is due to the fact that MEG3 will increase the expression of p53 and at the same time increase the level of nuclear factor expression κB (NF-κB) [69]. MEG3 represses the PI3K / AKT / mTOR pathway, this causes a decrease in the oncogenic behavior of miR- 21 that is overexpressed in breast cancer [13].
Development of lncRNAs-Based Cancer Therapies
Based on these studies, new therapies have been designed that involve the study of lncRNAs and their involvement in cancer, and several strategies have been implemented to cancer therapy using lncRNA, one strategy that has been implemented is to re-introduce a wild type lncNA with tumor suppressor function to cells that lack it. Another implemented strategy is the use of siRNAs, antisense oligonucleotides (ASOs) and viral vectors containing short-tip RNAs (shRNAs), which are involved in the expression of oncogenic lncRNAs. As a result of ASO-mediated inhibition of lncRNAs in animal models has been likely (Figure 2) [53].
Studies have shown that inhibition of the HOTAIR oncogene could reverse the EMT process and counter cell proliferation in breast cancer [32]. It was found that siRNA-mediated silencing of oncogenic lymphocytes has led to decreased viability of prostate cancer cells, the functioning of which begins with siRNA mediated by PRNCR1 knockdown, this silencing appears to be practical in inhibiting cancer cell proliferation and metastasis ability (Figure 2a) [53].
Another strategy for cancer therapy is to produce a splice variant, which is done by altering splice events. The application of lncRNA has also been sought, this application is done by inhibition of the interaction of lncRNAs with their objectives. This relationship has been proven with the inhibition of interactions between HOTAIR and PRC2 or LSD1 complexes. When this interaction is inhibited, the metastatic potential of cells in breast cancer is minimized [70].
Recent studies indicate that CRISPR / Cas9 is a useful tool for silencing transcription in loci expressed by some lncRNA (Figure 2b) [71,72]. The use of CRISPR/Cas9 served as a signaling of the transcriptional site of a gene promoter involved in transcription silencing [73]. Studies have shown that more than 16,000 lncRNA promoters could target human genome RNA [74]. CRISP/Cas9 has been used to generate an attack on the genomic DNA of cancer cells, an example of which is the elimination of lncRNA-NEAT1 and lncRNA-MALAT1, this modification served as an inhibition on the metastasis of cancer cells [14,15].
ARA is an lncRNA that has been implicated in several signaling pathways that are involved in the development of breast cancer, with cell adhesion, MAPK and pathways that regulate cell cycle progression. The lncRNA transcriptome profile in adriamycin-resistant breast cancer cells revealed that ARA is directly involved in adriamycin resistance. Studies found that decreased expression of ARA is reflected in reversal of drug resistance, equally in inhibition of cell proliferation and migration and in promotion of apoptosis and arrest of G2/M in adriamycin-resistant cells [16].
In superior transfection RNA (RNAi) interference techniques, there are viral vectors that mainly include recombinant adenovirus, lentivirus and retrovirus vectors (Figure 2c). RNA neutralization is performed, this is directed by the exogenous double stranded RNA, the RNAi is a biological process that allows the specific elimination of genes [17-19]. It is a specific technique, but the efficiency of siRNA undergoes instability, stemshloRNA can provide a lasting effect in vivo [75-78]. For the treatment of cancer, the use of shRNA to generate an attack on lncRNA has been studied with great interest. The inactivation of lncRNA-BCAR4 in lentiviral transfection significantly inhibited the formation of metastases in breast cancer [79]. By transfecting HOTAIR mRNA with a retrovirus in a gastric cancer cell line, it led to the inhibition of cell propagation in peritoneal dissemination [80]. Some lncRNA that have a suppressive function have a low expression in tumors, it is sought to increase the expression of these lncRNA to create a treatment for cancer. Virus transfection, as the main method for accurately transmitting the shRNA plasmid to the target site, can also be used to upregulate the corresponding lncRNAs by transfecting exogenously synthesized lncRNA plasmids into cancer cells [81].
Recent research has shown that LncRNAs are not junk DNA, rather they are a great tool for the study of cancer, causing research on the roles of lncRNA to take a new direction for research on tumorigenic mechanisms and cancer therapies.
There is great potential to develop biomarkers that help us create new therapies to treat breast cancer, as LncRNAs represent novel and rarely characterized components of cancer cells, this is because LncRNAs have been classified into oncogenic lncRNA and lncRNA suppressors of tumors.
Conflict of Interest
The authors declare no conflict of interest.
References
2. Perou CM, Sørlie T, Eisen MB, Van De Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge Ø. Molecular portraits of human breast tumours. Nature. 2000 Aug;406(6797):747-52.
3. Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, Zackrisson S, Cardoso F. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of Oncology. 2015 Sep 1;26(suppl_5):v8-30.
4. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010 Apr;464(7291):1071-6.
5. Silva JM, Boczek NJ, Berres MW, Ma X, Smith DI. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biology. 2011 May 1;8(3):496-505.
6. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009 Mar;458(7235):223-7.
7. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development. 2011 Sep 15;25(18):1915-27.
8. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007 Jun 29;129(7):1311-23.
9. Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012 Dec 14;338(6113):1435-9.
10. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Molecular Cancer. 2011 Dec;10(1):38.
11. Bach DH, Lee SK. Long noncoding RNAs in cancer cells. Cancer letters. 2018 Apr 10;419:152-66.
12. Zhou J, Zhou Y, Wang CX. LncRNA-MIAT regulates fibrosis in hypertrophic cardiomyopathy (HCM) by mediating the expression of miR-29a-3p. Journal of Cellular Biochemistry. 2019 May;120(5):7265-75.
13. Zhu M, Wang X, Gu Y, Wang F, Li L, Qiu X. MEG3 overexpression inhibits the tumorigenesis of breast cancer by downregulating miR-21 through the PI3K/Akt pathway. Archives of Biochemistry and Biophysics. 2019 Jan 1;661:22-30.
14. Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W, Leucci E. p53 induces formation of NEAT1 lncRNAcontaining paraspeckles that modulate replication stress response and chemosensitivity. Nature Medicine. 2016 Aug;22(8):861.
15. Mendell JT. Targeting a long noncoding RNA in breast cancer. New England Journal of Medicine. 2016 Jun 9;374(23):2287-9.
16. Jiang M, Huang O, Xie Z, Wu S, Zhang X, Shen A, Liu H, Chen X, Wu J, Lou Y, Mao Y. A novel long noncoding RNA-ARA: adriamycin resistance associated. Biochemical Pharmacology. 2014 Jan 15;87(2):254-83.
17. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998 Feb;391(6669):806-11.
18. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001 May;411(6836):494-8.
19. Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Current Opinion in Cell Biology. 2007 Jun 1;19(3):281-9.
20. Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, Mattick JS. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009 Nov 1;15(11):2013- 27.
21. Askarian-Amiri ME, Seyfoddin V, Smart CE, Wang J, Kim JE, Hansji H, Baguley BC, Finlay GJ, Leung EY.Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer. PloS One. 2014;9(7).
22. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz- Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007 Dec 21;318(5858):1917-20.
23. Zhang T, Hu H, Yan G, Wu T, Liu S, Chen W, et al. Long Non-Coding RNA and Breast Cancer. Technology in Cancer Research & Treatment, 2019 Apr 14;18:1533033819843889.
24. Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biology. 2013 Nov 1;14(11):R131.
25. Youness RA, Gad MZ. Long non-coding RNAs: functional regulatory players in breast cancer. Noncoding RNA Research. 2019 Feb 5.
26. Esmatabadi MJ, Motamedrad M, Sadeghizadeh M. Down-regulation of lncRNA, GAS5 decreases chemotherapeutic effect of dendrosomal curcumin (DNC) in breast cancer cells. Phytomedicine. 2018 Mar 15;42:56-65.
27. Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009 Jan;28(2):195-208.
28. Zhao W, Luo J, Jiao S. Comprehensive characterization of cancer subtype associated long noncoding RNAs and their clinical implications. Scientific Reports. 2014 Oct 13;4(1):1-7
29. Jiang M, Huang O, Xie Z, Wu S, Zhang X, Shen A, Liu H, Chen X, Wu J, Lou Y, Mao Y. A novel long noncoding RNA-ARA: adriamycin resistance associated. Biochemical Pharmacology. 2014 Jan 15;87(2):254-83.
30. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular Cell. 2010 Sep 24;39(6):925-38.
31. Gutschner T, Hämmerle M, Diederichs S. MALAT1—a paradigm for long noncoding RNA function in cancer. Journal of Molecular Medicine. 2013 Jul 1;91(7):791-801.
32. Gutschner T, Hämmerle M, Eißmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Groß M, Zörnig M. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer research. 2013 Feb 1;73(3):1180-9.
33. Wang L, Zeng X, Chen S, Ding L, Zhong J, Zhao JC, Wang L, Sarver A, Koller A, Zhi J, Ma Y. BRCA1 is a negative modulator of the PRC2 complex. The EMBO Journal. 2013 May 29;32(11):1584-97.
34. Gu J, Wang Y, Wang X, Zhou D, Shao C, Zhou M, He Z. Downregulation of lncRNA GAS5 confers tamoxifen resistance by activating miR-222 in breast cancer. Cancer Letters. 2018 Oct 10;434:1-0.
35. Conte F, Fiscon G, Chiara M, Colombo T, Farina L, Paci P. Role of the long non-coding RNA PVT1 in the dysregulation of the ceRNA-ceRNA network in human breast cancer. PLoS One. 2017;12(2).
36. Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. Cell cycle regulation by long non-coding RNAs. Cellular and Molecular Life Sciences. 2013 Dec 1;70(24):4785-94.
37. Khorshidi A, Dhaliwal P, Yang BB. Noncoding RNAs in tumor angiogenesis. InThe Long and Short Non- Coding RNAs in Cancer Biology 2016 (pp. 217-241). Springer, Singapore.
38. Beckedorff FC, Ayupe AC, Crocci-Souza R, Amaral MS, Nakaya HI, Soltys DT, Menck CF, Reis EM, Verjovski- Almeida S. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genetics. 2013 Aug;9(8).
39. Congrains A, Kamide K, Ohishi M, Rakugi H. ANRIL: molecular mechanisms and implications in human health. International Journal of Molecular Sciences. 2013 Jan;14(1):1278-92./a>
40. Folkersen L, Kyriakou T, Goel A, Peden J, Mälarstig A, Paulsson-Berne G, Hamsten A, Franco-Cereceda A, Gabrielsen A, Eriksson P. Relationship between CAD risk genotype in the chromosome 9p21 locus and gene expression. Identification of eight new ANRIL splice variants. PloS One. 2009 Nov 2;4(11):e7677.
41. Wan G, Mathur R, Hu X, Liu Y, Zhang X, Peng G, Lu X. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cellular Signalling. 2013 May 1;25(5):1086-95.
42. Chang YT, Lin TP, Tang JT, Campbell M, Luo YL, Lu SY, Yang CP, Cheng TY, Chang CH, Liu TT, Lin CH.HOTAIR is a REST-regulated lncRNA that promotes neuroendocrine differentiation in castration resistant prostate cancer. Cancer Letters. 2018 Oct 1;433:43-52./a>
43. Su X, Malouf GG, Chen Y, Zhang J, Yao H, Valero V, Weinstein JN, Spano JP, Meric-Bernstam F, Khayat D, Esteva FJ. Comprehensive analysis of long noncoding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014 Oct;5(20):9864.
44. Shore AN, Herschkowitz JI, Rosen JM. Noncoding RNAs involved in mammary gland development and tumorigenesis: there’sa long way to go. Journal of Mammary Gland Biology and Neoplasia. 2012 Mar 1;17(1):43-58.
45. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010 Apr;464(7291):1071-6.
46. Gabory A, Jammes H, Dandolo L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. BioEssays. 2010 Jun;32(6):473-80.
47. Feil R. Epigenetic asymmetry in the zygote and mammalian development. International Journal of Developmental Biology. 2009 Mar 17;53(2-3):191-201.
48. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature Reviews Genetics. 2002 Jun;3(6):415-28.
49. Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Research. 2002 Nov 15;62(22):6442-6.
50. Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, Coll J, Dugimont T, Curgy JJ. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis. 2002 Nov 1;23(11):1885-95.
51. Berteaux N, Lottin S, Monté D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. Journal of Biological Chemistry. 2005 Aug 19;280(33):29625-36.
52. Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, Penn LZ. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis.Cancer Research. 2006 May 15;66(10):5330-7.
53. Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Onco-gene. 2003; 22: 8031–41.
54. Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Research. 2012 Jun 1;40(11):5034-51
55. Sharif J, Muto M, Takebayashi SI, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007 Dec;450(7171):908-12.
56. Vicent GP, Nacht AS, Zaurin R, Font-Mateu J, Soronellas D, Le Dily F, Reyes D, Beato M. Unliganded progesterone receptor-mediated targeting of an RNAcontaining repressive complex silences a subset of hormone-inducible genes. Genes & Development. 2013 May 15;27(10):1179-97.
57. Cooper C, Guo J, Yan Y, Chooniedass-Kothari S, Hube F, Hamedani MK, Murphy LC, Myal Y, Leygue E. Increasing the relative expression of endogenous noncoding Steroid Receptor RNA Activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Research. 2009 Jul 1;37(13):4518-31.
58. Lanz RB, Chua SS, Barron N, Söder BM, DeMayo F, O’Malley BW. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Molecular and Cellular Biology. 2003 Oct 15;23(20):7163-76.
59. Silva JM, Boczek NJ, Berres MW, Ma X, Smith DI. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biology. 2011 May 1;8(3):496-505.
60. Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. TRENDS in Genetics. 2007 Dec 1;23(12):614-22.
61. Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, Mo YY. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death & Disease. 2014 Jan;5(1):e1008-.
62. Ai B, Kong X, Wang X, Zhang K, Yang X, Zhai J, Gao R, Qi Y, Wang J, Wang Z, Fang Y. LINC01355 suppresses breast cancer growth through FOXO3-mediated transcriptional repression of CCND1. Cell Death & Disease. 2019 Jun 26;10(7):1-3.
63. Pickard MR, Williams GT. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Cancer Research and Treatment. 2014 Jun 1;145(2):359-70.
64. Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, Ru K, Mercer TR, Thompson ER, Lakhani SR, Vargas AC. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011 May 1;17(5):878-91.
65. Hu P, Chu J, Wu Y, Sun L, Lv X, Zhu Y, Li J, Guo Q, Gong C, Liu B, Su S. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget. 2015 Oct 20;6(32):32410.
66. Binabaj MM, Bahrami A, Bahreyni A, Shafiee M, Rahmani F, Khazaei M, Soleimanpour S, Ghorbani E, Fiuji H, Ferns GA, Ryzhikov M. The prognostic value of long noncoding RNA MEG3 expression in the survival of patients with cancer: A meta-analysis. Journal of Cellular Biochemistry. 2018 Nov;119(11):9583-90.
67. Wu M, Huang Y, Chen T, Wang W, Yang S, Ye Z, Xi X. LncRNA MEG3 inhibits the progression of prostate cancer by modulating miR-9-5p/QKI-5 axis. Journal of Cellular and Molecular Medicine. 2019 Jan;23(1):29-38.
68. Zhou J, Zhou Y, Wang CX. LncRNA-MIAT regulates fibrosis in hypertrophic cardiomyopathy (HCM) by mediating the expression of miR-29a-3p. Journal of Cellular Biochemistry. 2019 May;120(5):7265-75.
69. Zhang Y, Wu J, Jing H, Huang G, Sun Z, Xu S. Long noncoding RNA MEG3 inhibits breast cancer growth via upregulating endoplasmic reticulum stress and activating NF-κB and p53. Journal of Cellular Biochemistry. 2019 Apr;120(4):6789-97.
70. Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Research. 2011 Jan 1;71(1):3-7.
71. Koch L. Functional genomics: screening for lncRNA function. Nature Reviews Genetics. 2017 Jan 3;18(2):70.
72. Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014 Oct 23;159(3):647-61.
73. Thakore PI, D’ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nature Methods. 2015 Dec;12(12):1143.
74. Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, Mandegar MA. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017 Jan 6;355(6320):eaah7111.
75. Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nature Biotechnology. 2011 Jan;29(1):79.
76. Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, Sinha N. An optimized microRNA backbone for effective single-copy RNAi. Cell Reports. 2013 Dec 26;5(6):1704- 13.
77. Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. The loop position of shRNAs and premiRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012 Nov 9;151(4):900-11.
78. Watanabe C, Cuellar TL, Haley B. Quantitative evaluation of first, second, and third generation hairpin systems reveals the limit of mammalian vector-based RNAi. RNA Biology. 2016 Jan 2;13(1):25-33.
79. Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH, Hung MC. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014 Nov 20;159(5):1110-25.
80. Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, Iijima K. Enhanced expression of long noncoding RNA HOTAIR is associated with the development of gastric cancer. PloS One. 2013;8(10).
81. Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. American Journal of Cancer Research. 2019;9(7):1354.