Abstract
Exosomes, a type of extracellular vesicles (EVs), are secreted from cells and taken up again by other cell types. They have attracted attention as new intercellular communication factors involved in various biological processes. Multivesicular bodies (MVBs) are intracellular organelles that are sources of exosomes. Recently, molecular regulatory mechanisms of EVs and MVBs mediated by ubiquitin and ubiquitin-like proteins (UBLs) have been elucidated. These include regulation of MVBs by deubiquitinases and neddylation, ubiquitination of human epidermal growth factor receptor 2 (Her2), a well-known target of cancer therapy, the relationship between UBL3 and cancer, and the relationship between the novel coronavirus infection, COVID-19, and EVs. In this manuscript, we present recent progress in the field. Based on this information, we discuss novel potential therapeutic strategies for exosome-related diseases.
Keywords
Ubiquitin-like protein, MVBs, Exosome, UBL3, SARS-CoV-2
Commentary
The multivesicular body (MVB), also called late endosome, is a subset of specialized endosomal compartments rich in intraluminal vesicles (ILVs). Multiple ILVs accumulate within MVBs [1,2]. ILVs are formed by invagination of the limiting membrane of early endosomes and budding into the lumen of the organelle. ILVs sequester specific proteins, lipids and cytosolic components. Although exosome release is known to be mediated by MVB, its regulation is not fully understood. Once MVBs fuse with lysosomes, the cargo of the ILVs is degraded. On the other hand, when MVBs fuse with the plasma membrane, the contents of ILVs are secreted outside the cell via exosomes. Most synthesized proteins are modified by post-translational modifiers, which regulate the amount, localization, stability, and activity of proteins. Post-translational modifications (PTM) are involved in the regulation of cellular functions [3]. The formation of MVB is known to be regulated by the endosomal sorting complexes required for transport (ESCRT) systems [1], as well as tetraspanins and UBLs. ESCRT systems are also dependent on ubiquitination [4]. Recently, ubiquitin and UBLs were reported to be involved in the regulation of ILV and MVB. Proteins modified by ubiquitin, SUMO, or UBL3 were incorporated into MVB.
Regarding the fusion of MVB to the plasma membrane and exosome release, the Rab family of small GTPase, including Rab27 [5], Rab11 [6], Rab35 [7], and cortactin [8], are responsible for the process. It was reported that Rab27 is ubiquitinated and degraded by the ubiquitin-proteasomal pathway and that the exosome release is regulated by inhibiting its ubiquitination [9]. In other words, KIBRA binds to Rab27a and inhibits the ubiquitination of Rab27, thereby increasing exosome release. Interestingly, KIBRA has been identified as a molecule that affects memory [10,11]. A genome-wide association study (GWAS) performed to analyze factors affecting human memory revealed that individuals with the KIBRA (rs17070145) T allele performed significantly better on multiple episodic memory tasks than those with the homozygous C allele [10]. This study used functional magnetic resonance imaging (fMRI) to analyze brain activation states and found differences between alleles in areas related to memory recall. Furthermore, a follow-up cohort study has also shown the involvement of the KIBRA T allele in memory performance [12]. Interestingly, in patients with Alzheimer's disease (AD), the KIBRA T allele has been reported to influence the risk of developing the disease [13,14]. Neurons also release exosomes and the propagation of malignant proteins via exosomes is involved in neurodegenerative diseases. The analysis of the regulatory mechanisms of KIBRA is expected to become an important area of research in neurodegenerative diseases related to exosome regulation.
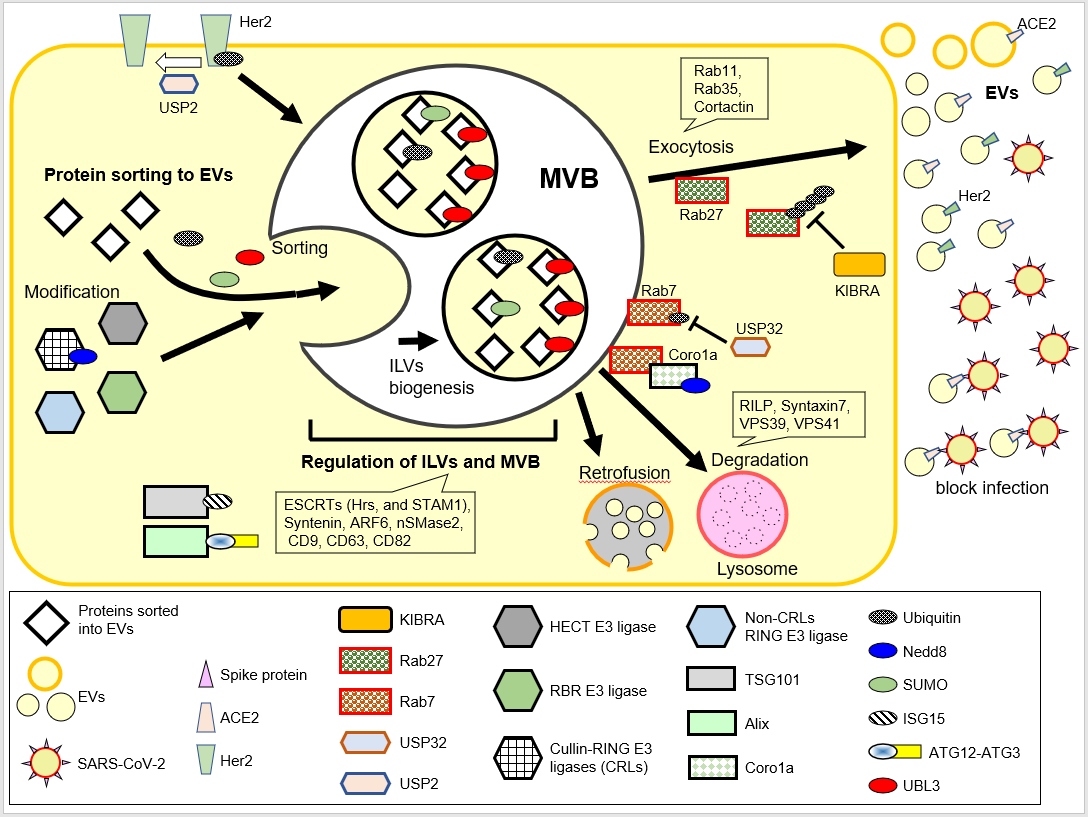
Figure 1: Schematic illustration of the regulation of MVBs and EVs by UBLs. UBLs (Ubiquitin, Nedd8, SUMO, ISG15, ATG12, and UBL3)are depicted as small oval shapes. The functions of UBLs have been described in our previous review [3]. In this review, we focus on the regulatory mechanisms of Her2 and ACE2 sorting, regulation of MVBs by Rab27 and Rab7, and retrofusion.
The ubiquitination level of target proteins is regulated by the balance between ubiquitination and deubiquitination. A search for deubiquitinases (DUBs) involved in the endosomal process, and MVB formation has led to the identification of ubiquitin-specific processing protease 32 (USP32) [4]. A loss of function in the gene encoding USP32 affects MVB size, content, and localization. Comprehensive proteomic analyses revealed that Rab7, the molecule that transports MVB to the lysosomal degradation pathway [15,16], is a target of USP32. Importantly, the amount of endogenous Rab7 was not altered by USP32 loss, indicating that Rab7 is not directly degraded by ubiquitination. Deubiquitination of Rab7 by USP32 is likely to promote the release of Rab7 from the membrane to the cytosol, which could affect MVB activity [4]. Furthermore, Coro1a was reported as a molecule that activates Rab7 [17]. Neddylation is a type of PTM that tags the ubiquitin-like protein NEDD8 to target proteins. Neddylated Coro1a binds to Rab7 and activates Rab7 [17]; therefore, MVBs are targeted by lysosomes for degradation, and exosome secretion is reduced. These results indicate that inhibitors of deubiquitination and neddylation enzymes have potential medical applications in exosome-related diseases. The crosstalk between these regulatory mechanisms, such as the mode of binding to Coro1a with ubiquitinated Rab7 and the biological effect of USP32 on Coro1a-bound Rab7, may be the next issue to be explored. The effects on KIBRA and DUBs mentioned above also seem to be of importance. Further analysis of these issues will help to elucidate the transport and sorting mechanisms of MVB and exosome release.
Recently, an interesting phenomenon was verified in the regulatory mechanism of MVBs. MVBs are known to have two fates: fusion with lysosomes, leading to the degradation of their contents, or fusion with the plasma membrane, leading to the release of their contents via exosomes. By combining a chemically induced dimerization (CID) system with microscopic analysis, the state of retrofusion, in which ILVs incorporated into MVBs are released back into the cytosol, was observed for the first time [18]. This retrofusion mechanism is believed to be involved in exosome uptake and major histocompatibility complex class II (MHC class II) antigen presentation. Interestingly, MVBs undergoing retrofusion are not involved in exosome release [18]. The retrofusion system may be involved in the mechanism of escape from the degradation of the virus after infection. Currently, it is not clear how the fate of the MVB is finely tuned. The relationship between the regulatory system of retrofusion and the roles of Rabs and UBLs is unknown. Further analysis is required to clarify the precise underlying mechanisms.
Exosomes are involved in a broad spectrum of biological phenomena and have been studied in various research fields. Her2 is a well-known marker of certain types of breast cancers [19] and is also a target of the anticancer drug Trastuzumab (clinical reagent also known as Herceptin) [20]. Interestingly, Her2 is known to be sorted into EVs [21, 22]. The regulatory system of sorting Her2 into EVs is important because Her2 on the EV surface inhibits Herceptin’s effect on cancer cells [23]. Her2 is also amplified in advanced and metastatic colorectal and gastric cancers [24]. Recently, it has been reported that Her2 is regulated by ubiquitin. Endogenous Her2 is ubiquitinated, and the IFN-γ-stimulated decrease in Her2 protein is inhibited by the knockdown of E3 ubiquitin ligase Cullin5 (CUL5) [25]. Another study showed that USP2 inhibitors promote Her2 ubiquitination and transport into the lysosome [26]. Although the effect of DUBs inhibitors on Her2 sorting into EVs is unknown, these results indicate that specific inhibitors of USP2 could be novel cancer drug candidates.
Our previous analysis reported that the novel posttranslational modification factor UBL3 regulates protein sorting into exosomes [27]. Recently, UBL3 has been identified as a cancer marker in studies comparing cancerous and normal tissues. Pancreatic cancer has a poor prognosis. Expression datasets of pancreatic cancer and normal pancreatic tissues were obtained from a database, and a comparative analysis identified CDKN2A, BRCA1, and UBL3 as variable molecules [28]. Interestingly, this is not the first report of an association between BRCA1 and UBL3. Although mutations in BRCA1/2 are known to induce breast cancer development, not all the breast cancer patients had mutations in BRCA1/2. A genome wide analysis of BRCA1/2 mutation-negative, high-risk breast cancer patients identified novel susceptibility loci for the PDE7B, UBL3, and CDKN2B-AS1 genes [29]. Therefore, these molecules may be involved in tumor progression. In addition, an analysis of gene expression between gastric cancer and normal tissue revealed that the expression levels of four genes (BGN, E2F3, PLAU, and SPP1) increased, and UBL3 gene expression levels decreased in gastric cancer and nonsmall cell lung cancer [30,31]. The effect of decreased UBL3 expression on EV release from gastric cancer cells and lung cancers is an issue. Since 60% of the molecules sorted into EVs are affected by UBL3, it is important to determine whether breast cancer-susceptible proteins, including Her2 and BRCA1, are molecules modified by UBL3.
SARS-CoV-2, the cause of the novel coronavirus infection, is transmitted by the binding of the spike protein on the virus surface to its target molecule, angiotensin-converting enzyme 2 (ACE2). Compared to seronegative controls, serum EVs from COVID-19 patients were found to contain high levels of ACE2 (evACE2). Furthermore, evACE2 showed 135-fold higher binding efficiency than vesicle-free recombinant human ACE2 (rhACE2) and 60-80-fold stronger ability to prevent SARSCoV- 2 infection [32]. The most important finding was that evACE2 was bound to the α, β, and δ SARS-CoV-2 variants. Thus, evACE2 is a potent inhibitor of novel variants that are expected to emerge in the future. evACE2-based therapeutic strategies are completely different from the available SARSCoV- 2 antiviral drugs and are therefore important as a means to combat novel viral variants that will emerge in the future. It has also been shown that ACE2 is degraded in lysosomes via ubiquitination by E3 ubiquitin ligase [33-35]. Therefore, ACE2 sorting into EVs is regulated by ubiquitination. From the viewpoint of pharmaceutical applications, it is important to search for the specific DUBs involved in this process. At present, whether the EV transport of ACE2 is regulated by ubiquitin or related UBLs is unknown. The search for agents that promote EV transport of ACE2 could become a completely different therapeutic strategy from the current SARS-CoV-2 therapeutics. It is expected to be a useful therapeutic agent against novel variants that will emerge in the future.
Since ubiquitination is involved in various biological phenomena, proteasome inhibitors have been used for medical applications. However, the lack of specificity of proteasome inhibitors is problematic. Therefore, inhibitors of specific E3 ubiquitin ligases have been developed [36]. Specific inhibitors of DUBs have also been explored [37]. Many recent studies have indicated that MVBs and exosome release are regulated by UBLs. The development of specific inhibitors will become increasingly important as novel medical strategies for exosome-related diseases in the future.
Acknowledgement
This work was supported by JSPS KAKENHI Grant Numbers JP19H05299, JP19H03427, JP21H00293, JP21K07159.
Conflict of Interest Disclosure
The authors have no conflicting financial interests.
References
2. Vietri M, Radulovic M, Stenmark H. The many functions of ESCRTs. Nat Rev Mol Cell Biol. 2020;21:25–42.
3. Ageta H, Tsuchida K. Post-translational modification and protein sorting to small extracellular vesicles including exosomes by ubiquitin and UBLs. Cell Mol Life Sci. 2019;76:4829–48.
4. Sapmaz A, Berlin I, Bos E, Wijdeven RH, Janssen H, Konietzny R, et al. USP32 regulates late endosomal transport and recycling through deubiquitylation of Rab7. Nat Commun. 2019;10:1454.
5. Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30.
6. Savina A, Fader CM, Damiani MT, Colombo MI. Rab11 promotes docking and fusion of multivesicular bodies in a calciumdependent manner. Traffic. 2005;6:131–43.
7. Hsu C, Morohashi Y, Yoshimura SI, Manrique-Hoyos N, Jung S, Lauterbach MA, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189:223–32.
8. Sinha S, Hoshino D, Hong NH, et al. Cortactin promotes exosome secretion by controlling branched actin dynamics. J Cell Biol. 2016;214:197–213.
9. Song L, Tang S, Han X, Jiang Z, Dong L, Liu C, et al. KIBRA controls exosome secretion via inhibiting the proteasomal degradation of Rab27a. Nat Commun. 2019;10:1639.
10. Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, et al. Common Kibra Alleles Are Associated with Human Memory Performance. Science. 2006;314:475–8.
11. Schneider A, Huentelman MJ, Kremerskothen J, Duning K, Spoelgen R, Nikolich K. KIBRA: a new gateway to learning and memory? Fronti Ag Neurosci. 2010;2:4.
12. Bates TC, Price JF, Harris SE, Marioni RE, Fowkes FG, Stewart MC, et al. Association of KIBRA and memory. Neurosci Lett. 2009;458:140–3.
13. Corneveaux JJ, Liang WS, Reiman EM, Webster JA, Myers AJ, Zismann VL, et al. Evidence for an association between KIBRA and late-onset Alzheimer’s disease. Neurobiol Aging. 2010;31:901–9.
14. Rodríguez-Rodríguez E, Infante J, Llorca J, Mateo I, Sánchez- Quintana C, García-Gorostiaga I, et al. Age-dependent association of KIBRA genetic variation and Alzheimer’s disease risk. Neurobiol Aging. 2009;30:322–4.
15. Vanlandingham PA, Ceresa BP. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem. 2009;284:12110–24.
16. Bucci C, Thomsen P, Nicoziani P, McCarthy J, Van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–80.
17. Fei X, Li Z, Yang D, Kong X, Lu X, Shen Y, et al. Neddylation of Coro1a determines the fate of multivesicular bodies and biogenesis of extracellular vesicles. Journal of Extracellular Vesicles. 2021; 10(12):e12153.
18. Perrin P, Janssen L, Janssen H, van den Broek B, Voortman LM, van Elsland D, et al. Retrofusion of intralumenal MVB membranes parallels viral infection and coexists with exosome release. Current Biology. 2021;31:3884-93.e4.
19. Lohrisch C, Piccart M. An overview of HER2. Semin Oncol. 2001;28:3–11.
20. Oh D-Y, Bang Y-J. HER2-targeted therapies - a role beyond breast cancer. Nat Rev Clin Oncol. 2020;17:33–48.
21. Koga K, Matsumoto K, Akiyoshi T, Kubo M, Yamanaka N, Tasaki A, et al. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res. 2005;25:3703–7.
22. Song X, Ding Y, Liu G, Yang X, Zhao R, Zhang Y, et al. Cancer Cell-derived Exosomes Induce Mitogen-activated Protein Kinasedependent Monocyte Survival by Transport of Functional Receptor Tyrosine Kinases. Journal of Biological Chemistry. 2016;291:8453– 64.
23. Marleau AM, Chen CS, Joyce JA, Tullis RH. Exosome removal as a therapeutic adjuvant in cancer. J Transl Med. 2012;10:134.
24. Nakamura Y, Okamoto W, Kato T, Esaki T, Kato K, Komatsu Y, et al. Circulating tumor DNA-guided treatment with pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer: a phase 2 trial. Nat Med. 2021;27:1899–903.
25. Jia Y, Kodumudi KN, Ramamoorthi G, Basu A, Snyder C, Wiener D, et al. Th1 cytokine interferon gamma improves response in HER2 breast cancer by modulating the ubiquitin proteasomal pathway. Molecular Therapy. 2021;29:1541–56.
26. Zhang J, Liu S, Li Q, Shi Y, Wu Y, Liu F, et al. The deubiquitylase USP2 maintains ErbB2 abundance via counteracting endocytic degradation and represents a therapeutic target in ErbB2-positive breast cancer. Cell Death Differ. 2020;27:2710–25.
27. Ageta H, Ageta-Ishihara N, Hitachi K, Karayel O, Onouchi T, Yamaguchi H, et al. UBL3 modification influences protein sorting to small extracellular vesicles. Nat Commun. 2018;9:3936.
28. Liu T, Chen L, Gao G, Liang X, Peng J, Zheng M, et al. Development of a Gene Risk Signature for Patients of Pancreatic Cancer. Journal of Healthcare Engineering. 2022;2022:1–8.
29. Lee JY, Kim J, Kim SW, Park SK, Ahn SH, Lee MH, et al. BRCA1/2- negative, high-risk breast cancers (BRCAX) for Asian women:genetic susceptibility loci and their potential impacts. Sci Rep. 2018;8:15263.
30. Shi Y, Qi L, Chen H, Zhang J, Guan Q, He J, et al. Identification of Genes Universally Differentially Expressed in Gastric Cancer. BioMed Research International. 2021;2021:1–9.
31. Zhao X, Yongchun Z, Qian H, Sanhui G, Jie L, Hong Y, et al. Identification of a potential tumor suppressor gene, UBL3, in nonsmall cell lung cancer. Cancer Biol Med. 2020;17:76–87.
32. El-Shennawy L, Hoffmann AD, Dashzeveg NK, McAndrews KM, Mehl PJ, Cornish D, Yu Z, et al. Circulating ACE2-expressing extracellular vesicles block broad strains of SARS-CoV-2. Nat Commun. 2022;13:405.
33. Deshotels MR, Xia H, Sriramula S, Lazartigues E, Filipeanu CM. Angiotensin II mediates angiotensin converting enzyme type 2 internalization and degradation through an angiotensin II type I receptor-dependent mechanism. Hypertension. 2014;64:1368–75.
34. Shen H, Zhang J, Wang C, Jain PP, Xiong M, Shi X, et al. MDM2-Mediated Ubiquitination of Angiotensin-Converting Enzyme 2 Contributes to the Development of Pulmonary Arterial Hypertension. Circulation. 2020;142:1190–204.
35. Wang G, Zhao Q, Zhang H, Liang F, Zhang C, Wang J, et al. Degradation of SARS-CoV-2 receptor ACE2 by the E3 ubiquitin ligase Skp2 in lung epithelial cells. Front Med. 2021;15:252–63.
36. Lydeard JR, Harper JW. Inhibitors for E3 ubiquitin ligases. Nat Biotechnol. 2010;28:682–4.
37. Schauer NJ, Magin RS, Liu X, Doherty LM, Buhrlage SJ. Advances in Discovering Deubiquitinating Enzyme (DUB) Inhibitors. J Med Chem. 2020;63:2731–50.
