Abstract
Innate immunity mediates anti-tumor responses through a variety of mechanisms including stimulation of cytokine production, activation of cytotoxic immune cells, and induction of cancer cell apoptosis. Together, these anti-tumor defense mechanisms can increase the efficacy of chemo- or immunotherapies. Intriguingly, recent evidence suggests that innate immune responses are intricately regulated not only by exogenous non-self RNA but also by host-derived RNAs such as pseudogene transcripts. Indeed, although pseudogenes have long been considered as non-functional artifacts of evolution, accumulating evidence indicates that pseudogene transcripts function as important gene expression regulators or immune modulators. In this article, we highlight recent findings that unveiled novel roles for pseudogene RNAs in antiviral or antitumor immunity, with a focus on the BRCA1 pseudogene transcripts that serve as immunoregulatory RNAs in breast cancer. Considering the importance of innate immune sensing and signaling in anti-tumor immunity, recent findings on the regulation of innate immunity by pseudogene RNAs may impact the design of next-generation antitumor therapies.
The Role of Antiviral Innate Responses in Tumor Immunity and Immunotherapy
Tumor immunity and immunotherapy have become increasingly important in treatment strategies for a variety of malignancies including advanced triple negative breast cancer [1,2]. Although immunotherapy has been shown to be effective, patient response rates vary significantly and only a small fraction of patients respond favorably to the treatment [3]. The efficacy of cancer immunotherapy appears to depend on the host immune system recognizing and eliminating cancer cells [4]. Increasing evidence demonstrates a positive correlation between the presence of host antitumor immune responses and favorable patient outcomes for many cancers [5-8]. As an example, tumors with a high density of tumor-infiltrating lymphoid cells (TILs) in the tumor microenvironment are more likely to respond to immune checkpoint inhibitors, whereas those with low or no TILs are less likely to respond to the inhibitors [9-11]. Thus, interventions that render nonresponding tumors to become responding tumors and hence promote antitumor immunity bear tremendous therapeutic potential.
Antiviral innate immune responses hold intrinsic anticancer benefits by promoting antitumor immunity and thereby increasing efficacy of chemotherapy and immunotherapy [4,12-16]. They mediate essential antitumor responses through several mechanisms, including stimulation of cytokine production, activation of cytotoxic immune cells, and induction of cancer cell apoptosis. Activation of innate immunity is triggered by pattern recognition receptors (PRRs) that are expressed in many different cells types (both immune and nonimmune cells) and detect invariant molecular structures shared by pathogens of various origins [17,18]. PRRs can be categorized into two subfamilies depending on their subcellular locations: the membrane-bound Toll-like receptors (TLRs) and the cytosolic nucleic acid sensors, such as RIG-I-like receptors (RLRs) and the cyclic GMPAMP synthase (cGAS)-stimulator of interferon gene (STING) pathway [19-22].
RIG-I and MDA5 of the RLR family are important PRRs involved in the detection of RNA viruses [19,20,23]. RLRs initiate host signaling that induces type I and III interferons (IFNs) and other cytokines, leading to the transcription of hundreds of IFN-stimulated genes (ISGs) [24]. Activation of RLRs by double-stranded RNA ligands not only triggers host immune responses but can also directly induce apoptosis of cancer cells, in an IFNdependent or independent manner [25]. Consequently, cancer cells are highly susceptible to RLR-induced cell death via intrinsic and extrinsic apoptosis and immune activation, indicating that the RLR signaling pathway is a promising molecular pathway to target in cancer immunotherapy. Thus, boosting innate immune responses may be key for establishing clinically desired antitumor immunity. Clinical trials evaluating the safety and efficacy of RLR agonists, STING agonists, TLR agonists, or poly(I:C) derivatives (synthetic analogues of doublestranded RNA) are currently completed or are ongoing [26-32]. Oncolytic viruses have also emerged as important agents in cancer treatment as they offer the attractive therapeutic combination of tumor-specific cell lysis and immune stimulation [4,22,33].
Regulation of Innate Antiviral Immunity by Host-derived RNAs
Interestingly, recent evidence suggests that antiviral innate immunity is regulated not only by exogenous nonself RNA but also by host-derived RNAs such as pseudogene transcripts. Pseudogenes have been considered nonfunctional artifacts of evolutionary processes due to degenerative features such as the accumulation of disruptive mutations or their lack of regulatory elements [34,35]. However, a growing body of evidence indicates a biological role for pseudogenes as gene expression regulators or immune modulators. According to the estimate of GENCODE [36], the human genome expresses 14,112 pseudogenes, a figure comparable to the number of protein-coding genes. The human genome also contains pseudogenes of tumor suppressors and oncogenes, including PTEN [37], KRAS [38], BRAF [39], p53 [40], and BRCA1 [41-43]. Pseudogenes of PTEN, KRAS, and BRAF were reported to regulate expression of their parent genes by sequestering microRNAs [44], or to function as competitive endogenous RNAs [45,46]. Other evidence indicates that pseudogenes may regulate gene expression by generating siRNAs [47,48] or by modulating RNA stability [44,49].
Pseudogene transcripts also serve as immune modulators. 5S ribosomal RNA pseudogene transcripts (in particular RNA5SP141) were shown to bind to RIG-I and induce the expression of antiviral or proinflammatory cytokines during infection with herpes simplex virus type 1 or the related herpesvirus Epstein-Barr virus [50].
During the course of infection with these viruses, specific RNA5SP141-binding proteins are downregulated, which leads to ‘unmasking’ of these pseudogene transcripts and thereby activation of RIG-I. A ribosomal protein S15a pseudogene transcript (RPS15AP4, Lethe) is selectively induced by proinflammatory cytokines or glucocorticoid receptor agonists, and serves as a functional regulator of inflammatory signaling through an interaction with NF- κB [51]. Many long noncoding RNAs (lncRNAs) are also known to function as positive or negative regulators of the innate antiviral response (as reviewed in detail elsewhere [52-54]). NEAT1 (nuclear paraspeckle assembly transcript 1) binds splicing factor proline and glutamine rich (SFPQ), which is required for the proper expression of several innate immune-related genes, and thereby increases antiviral gene induction (e.g. IL8, RIG-I and MDA5) [55]. THRIL (TNF-α and hnRNAPL related immunoregulatory lncRNA) interacts with hnRNPL at the TNF promoter and induces TNF-α transcription [56]. These data indicate that host-derived pseudogene RNAs and lncRNAs play pivotal roles in regulating antiviral defense and inflammatory signaling pathways.
The Presence of BRCA1 and BRCA1 Pseudogene (BRCA1P1) on Chromosome 17 in Breast Cancer
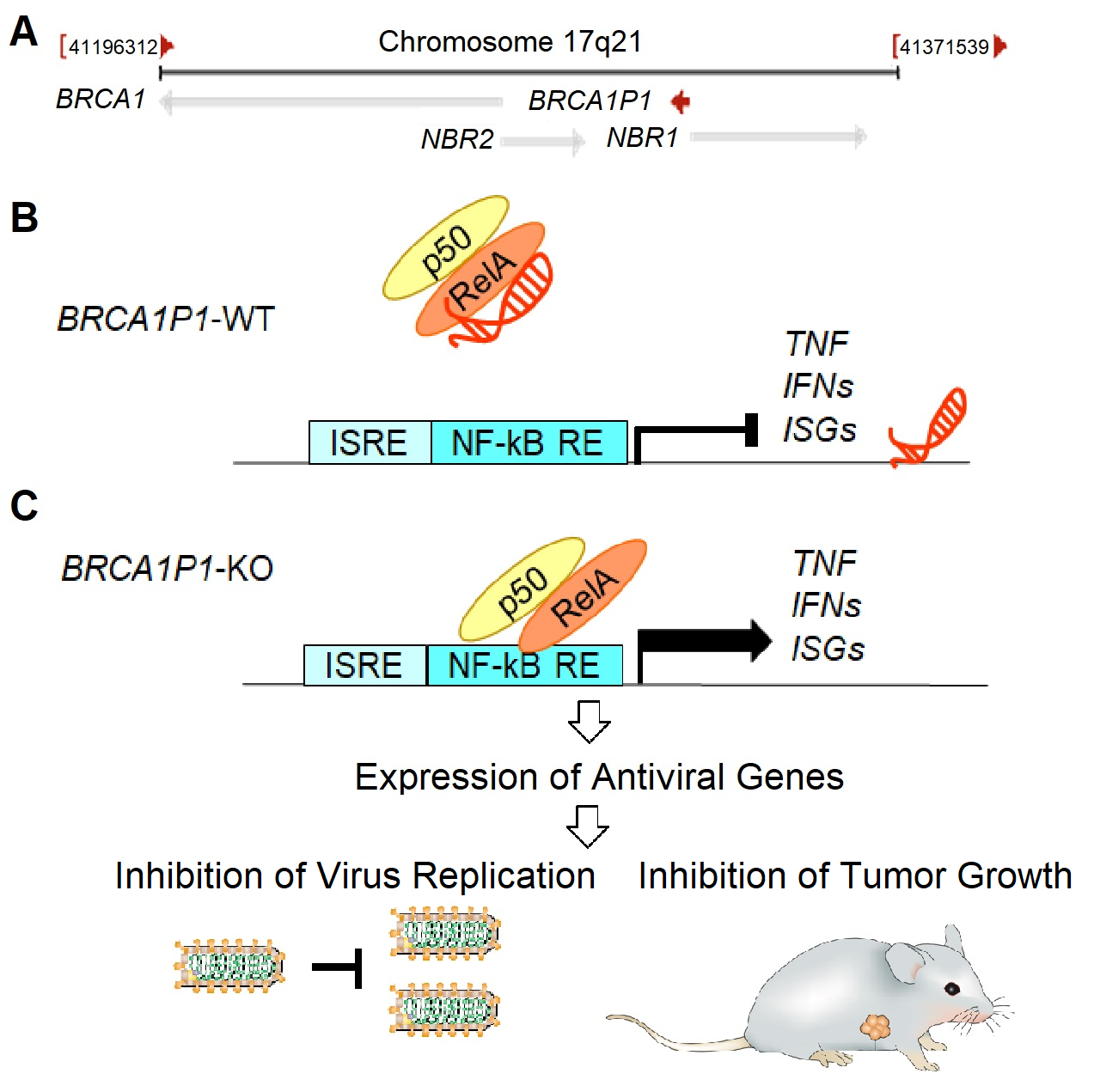
The chromosome 17q21 region that contains the BRCA1 gene has a partially duplicated pseudogene, BRCA1P1 (Figure 1A). It is of note that somatic abnormalities in chromosome 17 are common in breast tumors, including whole chromosome and gene-copy-number anomalies, allelic losses, and structural rearrangements [57-60]. These chromosome abnormalities have been linked to mechanisms of breast-cancer pathophysiology [61] associated with different clinicopathological features and gene-expression subtypes of breast cancer [62]. Abnormalities of specific loci on chromosome 17 including BRCA1 loss, P53 loss, ERBB amplification, and TOP2A amplification or deletion are known to have important roles in breast-cancer pathophysiology [63].
The BRCA1P1 pseudogene contains only three of the 24 exons of BRCA1 [42,43,64]. It also includes an insertion of the acidic ribosomal phosphoprotein P1 pseudogene (RPLP1P4) in exon 1a, and displays unique features of a chimeric pseudogene derived from two parent genes, BRCA1 and RPLP1. The presence of BRCA1P1 on the same chromosome close to BRCA1 appears to create a hotspot for homologous recombination, leading to genomic rearrangements between BRCA1P1 and BRCA1 in members of families with a high risk of breast cancer [65]. While the roles of BRCA1 in regulating homologous recombination and DNA damage repair have been studied extensively [66,67], the biological relevance of the BRCA1P1 pseudogene in breast cancer has not been elucidated. Recently, we discovered an important role for BRCA1P1 in regulating antiviral program-like responses in breast cancer cells [68]. In this article, we highlight recent findings unveiling a novel role for BRCA1P1 in antitumor immunity in breast cancer, and also discuss its potential impact for the design of next-generation anti-tumor therapies.
Regulation of Antiviral and Antitumor Immunity by the BRCA1P1 Pseudogene
We recently discovered a crucial role for the BRCA1P1 pseudogene in regulating antiviral and antitumor immunity in breast cancer [68]. BRCA1P1 expresses a lncRNA in the nuclei of breast cancer cells through divergent transcription using the bidirectional promoter between NBR1 and BRCA1P1 (Figure 1A). In the nuclei of BRCA1P1 wild type cells (BRCA1P1-WT), BRCA1P1-lncRNA binds to the NF- κB subunit RelA, inhibits the activity of RelA at its target promoters, and thereby negatively regulates transcription of antiviral genes (Figure 1B). In contrast, knockout of BRCA1P1 (BRCA1P1-KO) allows RelA to bind to its target promoters and to positively regulate transcription of antiviral genes, which increases antiviral-like host innate immune responses and suppresses viral replication in breast cancer cells (Figure 1C). In a xenograft mouse model of breast cancer, depletion of BRCA1P1 stimulates local immunity and suppresses tumor growth.
This discovery is of high significance in several aspects. First, this is the first study that demonstrates an important role for the BRCA1P1 pseudogene in antitumor responses through regulation of antiviral innate immunity and tumor growth. Second, while other pseudogene RNAs were shown to regulate antiviral or anti-inflammatory responses [51,69], our data derived from breast cancer cell lines and a breast cancer xenograft mouse model demonstrate a role for BRCA1P1 in both antiviral and antitumor immunity. Third, our results revealed that the effects of BRCA1P1 depletion are specific to cancer cells, with no induction of apoptosis in primary human mammary epithelial cells (HMEC). This suggests that normal cells may not experience toxicity from BRCAP1 inhibition, which induces cancer cell death specifically and immune cell activation through cytokine production. Fourth, as BRCA1P1-depleted cells were more sensitive to genotoxic drugs with increased apoptosis after doxorubicin and camptothecin treatment, our data suggest that BRCA1P1 depletion could be applied to increase chemotherapy sensitivity. Finally, as boosting innate immune responses is essential for effective antitumor immunotherapies, and since inhibition of BRCA1P1 robustly triggers the host innate immune system, there is therapeutic potential for BRCA1P1 depletion to increase sensitivity of immunotherapy, which might convert nonresponding tumors into responding tumors and hence promote the antitumor activity of immune checkpoint inhibitors.
Unique Features of BRCA1P1 and Future Studies
Although our study opens new possibilities to utilize antitumor immunity driven by BRCA1P1 depletion for cancer therapeutics, there are still limitations in our study. As we used athymic nude mice that lack T-cells, we were unable to evaluate the effects of BRCA1P1-deficiency on T-cells or other immune cells. Therefore, future studies using humanized mice will be needed to fully understand the relevance of BRCA1P1- lncRNA in modulating local immunity and the tumor microenvironment. Mechanistically, BRCA1P1-lncRNA binds to the NF-κB subunit RelA and inhibits the activity of RelA at its target promoters. Further investigation will be required to determine the mechanistic details of how BRCA1P1 regulates RelA activity, such as defining the RNA structural regions of BRCA1P1 that interact with RelA, by using chemical modifications and x-ray crystallography. Defining the RelA-interacting RNA motif may contribute to the design of therapeutic methods that specifically inhibit the interaction of BRCA1P1 with RelA.
It is of particular note that BRCA1P1 displays unique features of a chimeric pseudogene derived from two parent genes, BRCA1 and RPLP1. It contains processed sequences of RPLP1 from exon 1 to 3 (out of four exons of RPLP1) inserted in exon 1a of the BRCA1 gene. Compared to limited sequences originated from BRCA1 (three exons out of 24 exons of BRCA1), it contains the majority of the RPLP1 sequences, suggesting its role as a pseudogene transcript of ribosomal proteins. Given that RNA5SP141 and RPS15AP4 (Lethe) are also pseudogene transcripts originating from ribosomal RNA and ribosomal protein respectively [50,51], there might be a common role for ribosomal pseudogene transcripts in the regulation of innate immune pathways, which warrants further investigation. In terms of the genomic location of BRCA1P1, it is located in the chromosome 17q21 region close to BRCA1, which is frequently subject to somatic abnormalities in breast tumors [57-60]. Therefore, it is conceivable that BRCA1P1, along with BRCA1, may undergo genomic alterations during tumor evolution, which we will study further in the near future. Furthermore, it is worth noting that the BRCA1 and BRCA1P1 promoters have 85.7% identical sequences and are oriented in the same direction. As the two promoters have common cis-regulatory elements, they are likely regulated by similar transcription factors and upstream signals. Further studies will be required to identify the upstream signals that regulate BRCA1P1 expression, which would increase our understanding of the regulatory circuits in BRCA1P1-mediated immune responses.
Future Perspectives: Potential Relevance of Pseudogene RNAs and lncRNAs in Antiviral and Antitumor Therapy Development
Recent data from our and other groups demonstrated the importance of host-derived RNAs in the regulation of antiviral innate defense mechanisms. These studies suggested that the infected host cell has evolved to express cellular pseudogene RNAs or lncRNAs that counteract viral infection by stimulating antiviral gene induction. On the other hand, viruses may transcriptionally induce pseudogene RNAs or lncRNAs that dampen innate immune responses in order to facilitate virus replication. It is therefore tempting to speculate that the BRCA1P transcript may be one of the immune-dampening RNAs employed by viruses, which warrants future investigation. On the other hand, blocking the synthesis or function of host RNAs that negatively regulate the innate immune response may boost antiviral defense mechanisms, thereby facilitating virus clearance. Inhibition of these host-derived RNAs also may have therapeutic potential in cancer due to their abilities to stimulate antitumor immune responses and to increase sensitivity to chemotherapy and immunotherapy. Therefore, a molecular understanding of the regulation of innate immunity by host immunoregulatory RNAs may lead to the development of new clinical immunotherapies, which is an exciting area of future research.
Acknowledgments
We appreciate Lise Sveen’s careful editing of the manuscript. This work was supported by NCI P20CA233307 (OIO), Breast Cancer Research Foundation BCRF-20-071 (OIO and YH) and the Kiphart Global Health Research Fund (OIO). This study was further supported in part by NIH R01 AI087846 (MUG). OIO is an American Cancer Society Clinical Research Professor.
References
2. Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med. 2018;379:2108-21.
3. Sambi M, Bagheri L, Szewczuk MR. Current Challenges in Cancer Immunotherapy: Multimodal Approaches to Improve Efficacy and Patient Response Rates. J Oncol. 2019;2019:4508794.
4. Gujar S, Pol JG, Kim Y, Lee PW, Kroemer G. Antitumor Benefits of Antiviral Immunity: An Underappreciated Aspect of Oncolytic Virotherapies. Trends Immunol. 2018;39:209-21.
5. Pitt JM, Vetizou M, Daillere R, Roberti MP, Yamazaki T, Routy B, et al. Resistance Mechanisms to Immune- Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity. 2016;44:1255-69.
6. Kroemer G, Senovilla L, Galluzzi L, Andre F, Zitvogel L. Natural and therapy-induced immunosurveillance in breast cancer. Nat Med. 2015;21:1128-38.
7. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321-30.
8. Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest. 2015;125:3335-7.
9. Smyth MJ, Ngiow SF, Ribas A, Teng MW. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2016;13:143-58.
10. Fend L, Yamazaki T, Remy C, Fahrner C, Gantzer M, Nourtier V, et al. Immune Checkpoint Blockade, Immunogenic Chemotherapy or IFN-alpha Blockade Boost the Local and Abscopal Effects of Oncolytic Virotherapy. Cancer Res. 2017;77:4146-57.
11. Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17:559-72.
12. Bonaventura P, Shekarian T, Alcazer V, Valladeau- Guilemond J, Valsesia-Wittmann S, Amigorena S, et al. Cold Tumors: A Therapeutic Challenge for Immunotherapy. Front Immunol. 2019;10:168.
13. Newman JH, Chesson CB, Herzog NL, Bommareddy PK, Aspromonte SM, Pepe R, et al. Intratumoral injection of the seasonal flu shot converts immunologically cold tumors to hot and serves as an immunotherapy for cancer. Proc Natl Acad Sci U S A. 2020;117:1119-28.
14. Pyeon D, Vu L, Giacobbi NS, Westrich JA. The antiviral immune forces awaken in the cancer wars. PLoS Pathog. 2020;16:e1008814.
15. Simoni Y, Becht E, Fehlings M, Loh CY, Koo SL, Teng KWW, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575-9.
16. Zhao H, Ning S, Nolley R, Scicinski J, Oronsky B, Knox SJ, et al. The immunomodulatory anticancer agent, RRx- 001, induces an interferon response through epigenetic induction of viral mimicry. Clin Epigenetics. 2017;9:4.
17. Medzhitov R, Janeway CA, Jr. Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298-300.
18. Seth RB, Sun L, Chen ZJ. Antiviral innate immunity pathways. Cell Res. 2006;16:141-7.
19. Chan YK, Gack MU. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol. 2016;14:360-73.
20. Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20:537-51.
21. Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855-69.
22. Demaria O, Cornen S, Daeron M, Morel Y, Medzhitov R, Vivier E. Harnessing innate immunity in cancer therapy. Nature. 2019;574:45-56.
23. Liu G, Gack MU. Distinct and Orchestrated Functions of RNA Sensors in Innate Immunity. Immunity. 2020;53:26-42.
24. Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519-25.
25. Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. 2014;20:1301-9.
26. Brody JD, Ai WZ, Czerwinski DK, Torchia JA, Levy M, Advani RH, et al. In situ vaccination with a TLR9 agonist induces systemic lymphoma regression: a phase I/II study. J Clin Oncol. 2010;28:4324-32.
27. Demaria O, De Gassart A, Coso S, Gestermann N, Di Domizio J, Flatz L, et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc Natl Acad Sci U S A. 2015;112:15408-13.
28. Dowling DJ, Tan Z, Prokopowicz ZM, Palmer CD, Matthews MA, Dietsch GN, et al. The ultra-potent and selective TLR8 agonist VTX-294 activates human newborn and adult leukocytes. PLoS One. 2013;8:e58164.
29. Caskey M, Lefebvre F, Filali-Mouhim A, Cameron MJ, Goulet JP, Haddad EK, et al. Synthetic double-stranded RNA induces innate immune responses similar to a live viral vaccine in humans. J Exp Med. 2011;208:2357-66.
30. Ng KW, Marshall EA, Bell JC, Lam WL. cGAS-STING and Cancer: Dichotomous Roles in Tumor Immunity and Development. Trends Immunol. 2018;39:44-54.
31. Elion DL, Cook RS. Harnessing RIG-I and intrinsic immunity in the tumor microenvironment for therapeutic cancer treatment. Oncotarget. 2018;9:29007-17.
32. Iurescia S, Fioretti D, Rinaldi M. The Innate Immune Signalling Pathways: Turning RIG-I Sensor Activation Against Cancer. Cancers (Basel). 2020;12.
33. Gujar S, Pol JG, Kim Y, Lee PW, Kroemer G. Antitumor Benefits of Antiviral Immunity: An Underappreciated Aspect of Oncolytic Virotherapies. Trends Immunol. 2018;39:209-21.
34. Balakirev ES, Ayala FJ. Pseudogenes: are they “junk” or functional DNA? Annu Rev Genet 2003;37:123-51
35. Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541-58.
36. Pei B, Sisu C, Frankish A, Howald C, Habegger L, Mu X, et al. The GENCODE pseudogene resource. Genome Biol. 2012;13:R51.
37. Dahia PL, FitzGerald MG, Zhang X, Marsh DJ, Zheng Z, Pietsch T, et al. A highly conserved processed PTEN pseudogene is located on chromosome band 9p21. Oncogene. 1998;16:2403-6.
38. McGrath JP, Capon DJ, Smith DH, Chen EY, Seeburg PH, Goeddel DV, et al. Structure and organization of the human Ki-ras proto-oncogene and a related processed pseudogene. Nature. 1983;304:501-6.
39. Eychene A, Barnier JV, Apiou F, Dutrillaux B, Calothy G. Chromosomal assignment of two human B-raf(Rmil) proto-oncogene loci: B-raf-1 encoding the p94Braf/ Rmil and B-raf-2, a processed pseudogene. Oncogene. 1992;7:1657-60.
40. Zakut-Houri R, Oren M, Bienz B, Lavie V, Hazum S, Givol D. A single gene and a pseudogene for the cellular tumour antigen p53. Nature. 1983;306:594-7.
41. Smith TM, Lee MK, Szabo CI, Jerome N, McEuen M, Taylor M, et al. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 1996;6:1029-49.
42. Brown MA, Xu CF, Nicolai H, Griffiths B, Chambers JA, Black D, et al. The 5’ end of the BRCA1 gene lies within a duplicated region of human chromosome 17q21. Oncogene. 1996;12:2507-13.
43. Barker DF, Liu X, Almeida ER. The BRCA1 and 1A1.3B promoters are parallel elements of a genomic duplication at 17q21. Genomics. 1996;38:215-22.
44. Hirotsune S, Yoshida N, Chen A, Garrett L, Sugiyama F, Takahashi S, et al. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature. 2003;423:91-6.
45. Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344-57.
46. Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Leopold V, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319-32.
47. Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534-8.
48. Watanabe T, Totoki Y, Toyoda A, Kanedo M, Kuramochi-Miyagawa S, Obata Y, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539-43.
49. Han Y, Ma S, Yourek G, Park Y, Garcia J. A transcribed pseudogene of MYLK promotes cell proliferation. Faseb J. 2011;25:2305-12.
50. Chiang JJ, Sparrer KMJ, van Gent M, Lassig C, Huang T, Osterrieder N, et al. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat Immunol. 2018;19:53-62.
51. Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762.
52. Ahmed W, Liu ZF. Long Non-Coding RNAs: Novel Players in Regulation of Immune Response Upon Herpesvirus Infection. Front Immunol. 2018;9:761.
53. Fortes P, Morris KV. Long noncoding RNAs in viral infections. Virus Res. 2016;212:1-11.
54. Agliano F, Rathinam VA, Medvedev AE, Vanaja SK, Vella AT. Long Noncoding RNAs in Host-Pathogen Interactions. Trends Immunol. 2019;40:492-510.
55. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53:393-406.
56. Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111:1002-7
57. Nagai MA, Medeiros AC, Brentani MM, Brentani RR, Marques LA, Mazoyer S, et al. Five distinct deleted regions on chromosome 17 defining different subsets of human primary breast tumors. Oncology. 1995;52:448-53.
58. Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529-41.
59. Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, et al. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065-78.
60. Chin SF, Wang Y, Thorne NP, Teschendorff AE, Pinder SE, Vias M, et al. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007;26:1959-70.
61. Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96.
62. Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez- Boussard T, Lonning PE, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033-40.
63. Reinholz MM, Bruzek AK, Visscher DW, Lingle WL, Schroeder MJ, Perez EA, et al. Breast cancer and aneusomy 17: implications for carcinogenesis and therapeutic response. Lancet Oncol. 2009;10:267-77.
64. Pettigrew CA, French JD, Saunus JM, Edwards SL, Sauer AV, Smart CE, et al. Identification and functional analysis of novel BRCA1 transcripts, including mouse BRCA1-Iris and human pseudo-BRCA1. Breast Cancer Res Treat. 2010;119:239-47.
65. Puget N, Gad S, Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, et al. Distinct BRCA1 rearrangements involving the BRCA1 pseudogene suggest the existence of a recombination hot spot. Am J Hum Genet. 2002;70:858-65.
66. Legler K, Hauser C, Egberts JH, Willms A, Heneweer C, Boretius S, et al. The novel TRAIL-receptor agonist APG350 exerts superior therapeutic activity in pancreatic cancer cells. Cell Death Dis 2018;9:445
67. McLeod HL. Pharmacogenetics: more than skin deep. Nat Genet. 2001;29:247-8.
68. Han YJ, Zhang J, Lee JH, Mason JM, Karginova O, Yoshimatsu TF, et al. The BRCA1 Pseudogene Negatively Regulates Antitumor Responses through Inhibition of Innate Immune Defense Mechanisms. Cancer Res. 2021;81:1540-51.
69. Carnero E, Barriocanal M, Segura V, Guruceaga E, Prior C, Borner K, et al. Type I Interferon Regulates the Expression of Long Non-Coding RNAs. Front Immunol. 2014;5:548.