Abstract
Peripheral blood samples from patients diagnosed as HIV-1 positive and treated with anti-retroviral therapy (ART) for prolonged periods of time can present difficulties in current quantitative HIV-1 DNA detection, based on PCR assays. The current gold standard platform for HIV-1 DNA detection is real-time PCR, but its sensitivity is not always enough to detect extremely low levels of HIV- 1 DNA in some patients. We have developed a novel end-point PCR assay based on precision image pi-code (pCode) MicroDiscs detection platform. The newly developed “pCode assay” showed 100% sensitivity, while the real-time PCR assay showed 92.3%, in a head-to-head comparison using blood samples from 39 HIV-1 infected clinic patients, with a stable fully suppressed plasma viral-load (<20 copies/mL) for more than 2 years. Further analysis revealed a detection sensitivity over 27 times higher for the “pCode assay” compared to real-time PCR. The pCode assay is very specific and reproducible, with less than 20% Coefficient of Variation (CV) in detection, ranging from 40 to 4000 HIV-1 DNA copies per one million white blood cells. The pCode assay may have a great potential to be a standard assay platform in the future for the HIV diagnostic field, focusing on HIV cure trials.
Keywords
IV-1 DNA detection, Real-time PCR, End-point PCR
Abbreviations
HIV: Human Immune Deficiency Virus; HCV: Hepatitis C Virus
Introduction
Currently, around 60-75% of the HIV positive patients in developed countries on anti-retroviral therapy (ART) have undetectable plasma viral load using current diagnostic PCR assays [1,2]. However, most patients have been found in many research studies to maintain a detectable level of HIV DNA-infected lymphocytes, and if treatment is interrupted, plasma viral load returns to detectable levels within a few weeks [3,4]. Therefore, many current researches aim to study the possibility of therapeutic strategies to reduce this latent reservoir of HIV DNA positive cells [5,6].
Consistent detection of the presence and quantification of HIV-1 DNA in patient samples is one of the most technically challenging processes in the field of molecular diagnostics. It may be even more difficult to identify the presence of HIV-1 DNA when HIV-1 patients are under successful ART for a prolonged period [7-10].
Long duration of ART results in a reduction of HIV-1 DNA in peripheral blood mononuclear cells (PBMCs) [11-15]. It is estimated that HIV-1 infected cells are present in only 40-100 cells per one million of white blood cells [7-9,16-18], particularly if they commence ART with integrase inhibitor in early HIV-1 infection [19-21].
Quantification of HIV-1 DNA levels is important to evaluate the efficacy of ART in HIV-1 patients, since it has been widely reported that monitoring HIV- 1 DNA levels is associated with long-term efficacy of ART in HIV-1 infected individuals [6,11,22-31]. Currently, most clinical molecular laboratories use the TaqMan probe based real-time PCR detection method for quantification of HIV-1 DNA. There is one commercially available diagnostic assay offering HIV- 1 DNA detection, GeneXpert (Cepheid), which is also based on real-time PCR with TaqMan probe detection, providing qualitative analysis data [32-34].
In general, both, clinical diagnostic and molecular research laboratories, use real-time PCR as a gold standard method in molecular detection tests. Several reports have suggested that the real-time PCR method might not show the highest sensitivity [35,36]. Alternative detection methods have been evaluated to achieve higher sensitivity in several clinical sample analyses. These are based on end-point PCR detection method. A detection technique called enzymatic amplified detection of PCR amplified DNAs (“Enzyme- Linked Immunosorbent Assay (ELISA)-PCR”) has been evaluated to have better sensitivity than realtime PCR method. This approach combines PCR based amplification of targeted DNA, followed by enzymatic signal amplification detection, which has been used in EIA (Enzyme Immunoassay) with microtiter plate for several proteins [36-41]. Furthermore, the droplet digital PCR (ddPCR) method has been developed to achieve superior sensitivity and quantification capability to real-time PCR method. The ddPCR is also based on end-point PCR platform [9,28,42,43].
We have evaluated a totally new end-point PCR technique, developed by PlexBio. This detection technique is based on precision image MicroDisc detection, using an automated IntelliPlex1000 πCode processor machine based on circular πCode MicroDisc for detecting PCR amplified DNAs. The πCode endpoint PCR assay was initially developed for detection and identification of various oncogene markers and HCV genotyping [44]. This assay, however, was not systematically evaluated to assess detection sensitivity compared to gold standard real-time PCR assay.
In this report, we aimed to compare real-time PCR assays with the πCode end-point PCR detection assay for HIV-1 DNA. We used the well-characterized primer set of sk145-skcc1b for detecting HIV-1 gag region for this comparison. This primer set has been evaluated in an enormous number of articles in the past and was used in the diagnostic kit of the “Amplicor HIV- 1 detection assay” (Roche Diagnostic) [40,45,46], the first FDA approved HIV-1 DNA diagnostic assay. Since there is no commercial HIV-1 DNA assay available, we developed a quantitative real-time PCR assay with TaqMan probe using the sk145-skcc1b primer set, which have been extensively used in our previous HIV clinical trial studies [47-55] and basic studies on HIV-1 gene silencing and microRNA analysis [54,56-63]. We used our real-time PCR assay as a gold standard realtime PCR assay. We conducted several comparison experiments of real-time PCR assay and the πCode end-point assay in order to evaluate sensitivity and specificity.
Subjects and Methods
Study design, white blood cells preparation and ethics
Thirty-nine HIV-1-infected individuals, who had received ART, and showed completely suppressed levels of HIV-1 viral load (<20 copies/mL: HIV-1 RNA copy number in plasma, COBAS AmpliPrep/COBAS TaqMan HIV-1 Test) for more than 2 years, were analyzed in this study. The samples were from the NSW State Reference laboratory for HIV, St. Vincent’s Hospital (Darlinghurst, Australia). Demographics and HIV disease characteristics are shown in Supplementary Table 1. The white blood cells (WBCs) were obtained from 3 mL of whole blood in ACD (citric acid dextrose) tubes using the red blood cell lysis buffer (Sigma- Aldrich, 11814389001 Roche), following manufacturer’s instructions. The WBCs were counted using the C-Chip disposable Hemocytometer, (Cat # DHC-NO1-250, Nanoentek, Seoul, Korea), and the cell count number was used for normalization of HIV-1 DNA copy number. This study was approved by the St. Vincent’s Hospital Human Research Ethics Committee (HREC LNR/16/ SVH/327).
DNA extraction and real time PCR amplification
DNA extraction from WBCs was conducted with the Maxwell RSC system, an automated extraction instrument (Promega, Madison, WI, USA), using Maxwell RSC Buffy Coat DNA kit, Cat # AS1540 (Promega, Madison, WI, USA). Real-time PCR amplification was performed using forward primer of the sk145: 5′-AGT GGG GGG ACA TCA AGC AGC CAT GCA AAT-3′, and reverse primer of the skcc1b: 5′-TAC TAG TAG TTC CTG CTA TGT CAC TTC C-3′, and the LNA TaqMan probe: FAM 5′-ATC AAT GAG GAA GCT GC-3′ BHQ1, where LNA (locked nucleic acid) modified bases are shown with underline. Premix Ex Taq Probe qPCR kit (Cat # RR390W, Takara) was used with the LightCycler 480 real-time PCR machine (Roche). Reaction conditions were 95°C for 30 seconds as an initial denaturation of DNA, followed by 50 PCR cycles of 95°C for 7 seconds and 60°C for 30 seconds, using 0.25 μM of both primers and 0.075 μM of LNA TaqMan probe. A total volume of 50 μL per reaction, using 8 μL of extracted DNA, resulting in around 200,000 equivalent WBCs analyzed in each PCR reaction. Quantification of HIV-1 copy number per patient sample was determined from a standard curve generated with HIV-1 plasmid controls, with concentrations of 4,40,400, 4000 HIV- 1 copies/μL. The HIV-1 copy number for each sample, average value of duplicate reactions, was normalized with the cell count number. HIV-1 copy number per one million of WBCs was used as a standardized unit.
The πCode end-point PCR assay
Two πCode probes were designed for the πCode assay: sk-πCode set-1 probe: AmC6-5′-CAT CAA TGA GGA AGC TG-3′ and sk-πCode set-2 probe: AmC6-5′-AAG CTG CAG AAT GGG-3′, where “Am” indicates an amine modification at the 5′ end of the oligonucleotide and “C6” indicates a carbon six linker. A blank πCode probe was also prepared: Blank AMC6: AmC6-5′-AAT ATA ATA TAT TAT-3′. The blank probe was used to check the levels of non-specific binding signal for each “πCode end-point PCR assay” run. Coupling reactions were conducted using the pre-activated πCode MicroDiscs (PlexBio) and amine-modified probes according to the manufacturer’s instructions. After coupling reactions were completed, the covalently linked sk-πCode set-1 probe, the sk-πCode set-2 probe and the blank probe were prepared at a concentration of 100 MicroDiscs per μL in the πCode coupling storage buffer (PlexBio) and stored at 4°C until use.
Primers were the same as the real-time PCR, except that a biotin labeled reverse primer of skcc1b was used for the πCode assay. Like the real-time PCR analysis, a volume of 8 μL of extracted DNA sample was used, in duplicate, in the πCode end-point PCR assay. The reaction conditions were the same without the addition of the LNA TaqMan probe, and the πCode PCR was stopped at 35 cycles. The PCR was followed by hybridization and washing.
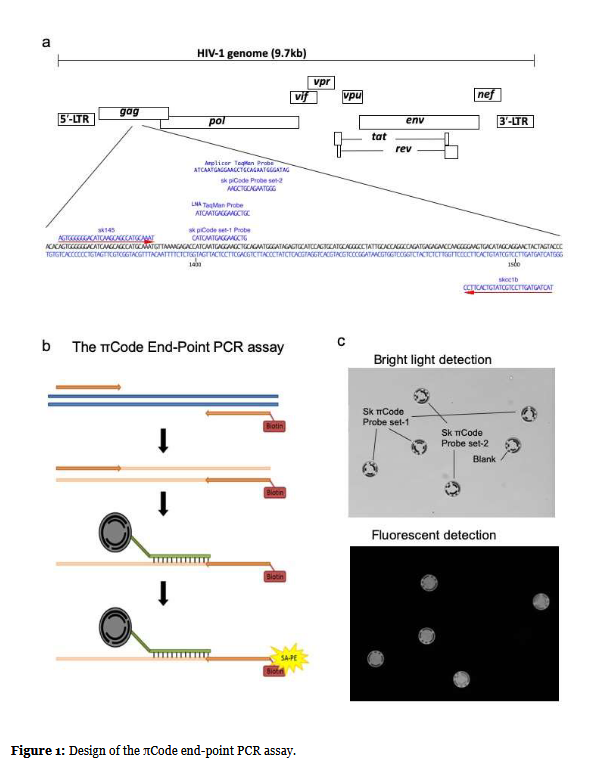
The hybridization of the amplified DNA product was performed in a 96-well microtiter plate, with a mix of 50 MicroDiscs of the three πCode probes (sk-πCode set- 1, sk-πCode set-2, and the blank probes). The program was 40°C for 20 minutes, followed by three times washing step with the washing buffer (PlexBio). After washing the πCode magnetic MicroDiscs, 50 μL of the fluorescent reagent of streptavidin-phycoerythrin (SAPE: PlexBio) was added to the reaction well, followed by four-times washing steps. All these reaction steps, including hybridization, washing, and adding of SAPE solution were performed in an automated machine, “IntelliPlex1000 πCode Processor”. The fluorescent signal generated from the πCode MicroDiscs was detected using the “PlexBio100 Analyzer”, equipped with two detection units. The machine uses a CCD camera to read the distinct image patterns of the MicroDiscs under bright field to identify the three probes. The analyzer also measured and calculated the mean fluorescence signal intensity (MFI) corresponding to the sk-πCode set-1 probe, the sk-πCode set-2 probe, and the blank probe, identified in the bright field (see the representative image in Figure 1c).
Quantification capability of the πCode end-point PCR assay
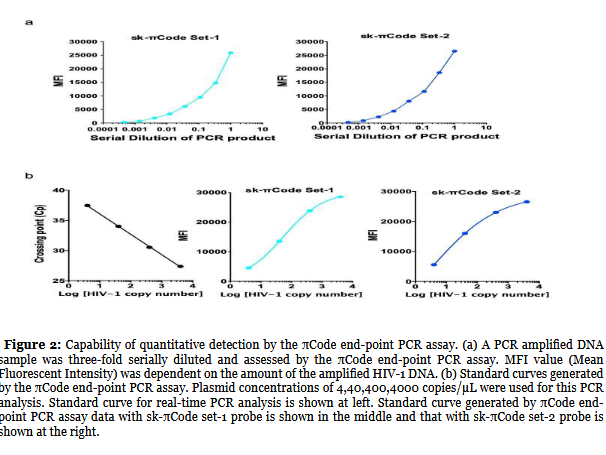
HIV-1 plasmid DNA at a concentration of 4000 HIV- 1 copies/μL was amplified by 50 PCR cycles with sk145 and skcc1b primer set, followed by three times dilution series of the PCR amplified DNA. The serially diluted DNAs were analyzed using IntelliPlex-1000 πCode Processor and PlexBio-100 Analyzer. Plasmid concentrations of 4,40,400,4000 HIV-1 copies/μL were used to assess the ability to do quantitative detection of known HIV-1 plasmid concentrations.
Identification of HIV-1 subtypes
To confirm HIV-1 subtype, protease/reverse transcriptase (RT/POL) regions were amplified with the in-house Drug Resistance assay, at the NSW State Reference laboratory for HIV, St Vincent’s Hospital, as described previously [49]. Briefly, DNA extracted from white blood cells was amplified by 1-step RT-PCR with Platinum Taq High Fidelity PCR kit (Invitrogen) for 30 minutes at 52°C, 2 minutes at 94°C, and 50 cycles of 15 seconds at 94°C, 30 seconds at 55°C, 1 min 30 seconds at 68°C using forward primer: 5′- TGA TGA CAG CAT GYC ARG GAG T -3′ and reverse primer: 5′- CTG CTA TTA ADT CTT TTG CTG GG -3′. In the second step, a nested PCR was performed with VELOCITY DNA Polymerase PCR kit (Bioline) using forward primer: 5′- GAA GGA CAC CAA ATG AAA GAY TG -3′ and reverse primer: 5′- GTA TGT CAT TGA CAG TCC AGC -3′ for 2 minutes at 98°C and 40 cycles of 30 seconds at 98°C, 30 seconds at 55°C, 1 minute 30 seconds at 72°C. The amplified DNA was subjected to a standard BigDye sequence analysis with 7 sequencing primers: Primer-1: 5′- GAA GGA CAC CAA ATG AAA GAY TG -3′, Primer-2: 5′- ATT GTT TAA CYT TTG GGC CAT CC -3′, Primer-3: 5′- TAG GAC CTA CAC CTG TCA ACA TAA TTG G -3′, Primer-4: 5′- CCA AAA GTT AAA CAA TGG CCA TTG ACA GA -39, Primer-5: 59- TCT AAA AGG CTC TAA GAT TTT TGT CAT GC -3′, Primer-6: 5′- GCT TCC ACA GGG ATG GAA AGG -3′, and Primer-7: 5′- CAG CAC TAT AGG CTG TAC TGT CCA -3′. HIV-1 subtype was identified using the HIV Drug Resistance Database, Stanford University.
Detection sensitivity comparison
A “head-to-head” comparison of detection sensitivity of both assays, real-time PCR and πCode end-point PCR, was conducted with HIV-1 DNA extractions from three random clinical samples. The samples were threefold serially diluted and evaluated with both assays. For further comparisons of these two assay formats, the three serially diluted samples were also analyzed with 50 cycles of the πCode end-point PCR assay, equivalent to the realtime PCR analysis.
Reproducibility of the πCode end-point PCR
Five clinical samples containing different amounts of HIV-1 DNA were tested. The experiment was independently repeated three times to obtain coefficient of variation (CV) values and assess reproducibility.
Detection specificity
The extracted DNA obtained from a HIV-2 sample was used for evaluating assay specificity of the πCode end-point PCR assay and the real-time PCR assay. Five healthy donors’ samples were used as controls. The HIV-2 sample was confirmed with HIV-2 infection using serological screening assay, followed by our western blot confirmation assay (Cat No. 72252 New LAV Blot II Assay, Bio-Rad). HIV-2 DNA detection was confirmed according to the in-house HIV-2 DNA assay procedure at St Vincent’s Hospital. In brief, DNA extracted from white blood cells was amplified with Taq DNA polymerase from Premix Ex Taq-Probe qPCR (Cat # RR390W, Takara), using a forward primer: 5′- GAG CCC TGA GAG GTT CTC-3′ and a reverse primer: 5′- GGT YTT TAA GCA AGC AAG CGT GG -3′ with TaqMan probe: 5′-FAM- AGA GTC TAG CAG GGA ACA CCC AGG C-3′ BHQ1. The PCR was performed in a Light-Cycler 480 Real-Time PCR machine (Roche) for 30 seconds at 95°C as an initial denaturation of DNA, and 50 cycles of 95°C for 7 seconds and 60°C for 30 seconds, using 0.25 μM of both primers and 0.075 μM of the TaqMan probe in a total volume of 40 μL per reaction.
Statistical analysis
Standard curves from known concentrations of HIV-1 plasmid copy numbers were generated with GraphPad Prism v7 (GraphPad Software), using the sigmoidal analysis function. Correlation analysis of the πCode endpoint PCR assay and real-time PCR assay was performed using Pearson correlation test in GraphPad Prism v7, and further comparison was done with Bland-Altman plot analysis in GraphPad Prism v7. All data analyses were also performed using GraphPad Prism v7.
Results
The πCode end-point PCR assay
The position in the HIV-1 genome of all primers and probes used in this study are shown in Figure 1a. Two probes were designed for the πCode end-point PCR assay (sk-πCode set-1 probe and sk-πCode set-2 probe). The two probe positions relative to the original Roche HIV- 1 Amplicor TaqMan probe are shown in Figure 1a. The two probes are located within the sequence region of the original Roche HIV-1 Amplicor TaqMan probe. The locked nucleic acid (LNA) TaqMan Probe was used for the real-time PCR analysis. The sequence of the LNA TaqMan probe is nearly identical to the sk-πCode set-1 probe with only one base difference (Figure 1a).
(a) HIV-1 genome structure is shown with HIV-1 gag region alignment of the primers and the probe used in this study. The forward primer of sk145 and the reverse primer of skcc1b primers target HIV-1 gag region, which were indicated with red arrows along with HIV-1 sequence. Locations of TaqMan probe for real-time analysis and sk- πCode set-1, sk-πCode set-2 probes for πCode end-point assay are also shown along gag sequence. TaqMan probe and sk-πCode set-1 were designed to hybridize an almost identical region with only one base difference. The sk- πCode set-2 probe targets 12 bases downstream region of that of the sk-πCode set-1 probe site. The original TaqMan probe in Amplicor HIV-1 detection assay is indicated at the top. (b) The πCode end-point PCR assay principle and procedure (See detailed description in Materials and Methods). (c) Bright light field and fluorescent field image captured by PlexBio100 Analyzer. Bright light phase image identified district πCode magnetic MicroDiscs linked with sk-πCode set-1 probe and the sk-πCode set- 2 probe. The blank probe was used for checking any non-specific binding to the πCode disc. The fluorescent detection image was shown with the same field as in the bright light image.
A diagram of the πCode end-point PCR assay procedure is shown in Figure1b. The reverse primer of skcc1b was labeled with Biotin. After PCR amplification, all PCR amplified DNAs contained the Biotin label. In the next reaction step, the Biotin labeled amplified DNAs were hybridized with the sk-πCode set-1 probe and the sk- πCode set-2 probe, each probe was covalently linked to πCode magnetic MicroDiscs with distinct image patterns. After washing the πCode MicroDiscs, the streptavidin-phycoerythrin (SA-PE) fluorescent solution was added to the reaction well, which was then captured through the high affinity reaction between the biotin and streptavidin, resulting in quantitative fluorescence from the phycoerythrin bound to πCode discs in samples containing HIV-1 DNA. All these reaction steps, including hybridization, washing, and adding of SA-PE reagent, were performed by the “IntelliPlex1000 πCode Processer” automated machine, using a 96-well microtiter plate. The PlexBio100 Analyzer, equipped with two detection units, was used for signal detection and quantitation. This analyzer uses a CCD camera to read the distinct image patterns of the πCode MicroDiscs under bright field and is also able to measure and calculate the mean fluorescence signal intensity (MFI) corresponding to the sk-πCode set- 1 probe and the sk-πCode set-2 probe (Figure 1c).
Quantification of the πCode end-point PCR assay
As an initial evaluation step, we needed to assess whether the πCode end-point PCR assay could quantitatively detect a range of concentrations of PCR amplified DNA. The amplified HIV-1 plasmid DNA (initial concentration of 4000 HIV-1 copies/ul) was three times serially diluted and analyzed using IntelliPlex-1000 πCode Processor and PlexBio-100 Analyzer generating MFI values. The data based on the set-1 probe is shown on the left figure of Figure 2a and the data based on the set-2 probe is shown on the right figure of Figure 2a. As shown, the values in MFI increased according to the concentration of PCR amplified DNA, suggesting that the πCode end-point PCR assay can do quantitative analysis of PCR amplified DNA.
In the next step, assessed whether or not the πCode endpoint PCR assay was able to achieve quantitative detection of the known concentration of HIV-1 plasmids. Plasmid concentrations of 4,40,400,4000 HIV-1 copies/μL were used for this assessment. The results for real-time PCR are shown on the left figure of Figure 2b. Cp cycle numbers obtained from real-time PCR were inversely correlated to Log10 value of HIV-1 plasmid copy numbers, as expected. In the πCode end-point PCR assay, the standard curves were generated using MFI values obtained from the known concentrations of plasmid copy numbers. The values obtained with sk-πCode set-1 probe are shown in the middle figure of Figure 2b, and the values for sk- πCode set-2 probe are shown in the right figure of Figure 2b. The data strongly suggested that the πCode end-point PCR assay can quantify HIV-1 copy numbers of unknown test samples, like real-time PCR analysis.
Comparison of real-time PCR and the πCode end-point PCR with diagnostic laboratory HIV-1 positive specimens
The next step of comparison of real-time PCR and the πCode end-point PCR using clinical samples. As described above, the πCode end-point PCR assay quantified HIV- 1 copy number for patient samples using MFI values. A standard curve generated with HIV-1 plasmid controls, with concentrations of 4,40,400,4000 HIV-1 copies/μL, was used for the estimation of HIV-1 copy number of the unknown samples. The data from 39 patient samples analyzed with the πCode end-point PCR using sk-πCode set-1 probe and set-2 probe, and the real-time PCR using TaqMan probe, are compared in Table 1. The HIV-1 copy numbers obtained from πCode end-point PCR using the sk-πCode set-1 probe and from real-time PCR showed similar values, except for ID No. 7,20, and 24, which contained very low levels of HIV-1 DNA and the real-time PCR was not able to detect.
Comparing HIV-1 copy numbers obtained from the πCode assay using sk-πCode set-1 probe with sk-πCode set- 2 probe, both probes showed similar values in general. However, in five samples (ID No. 3,6,10,18, and 35), the HIV-1 DNA levels using the sk-πCode set-2 probe showed less than one tenth of the HIV-1 DNA levels using the sk- πCode set-1 probe.
Correlation analysis to compare πCode end-point PCR and real-time PCR assays
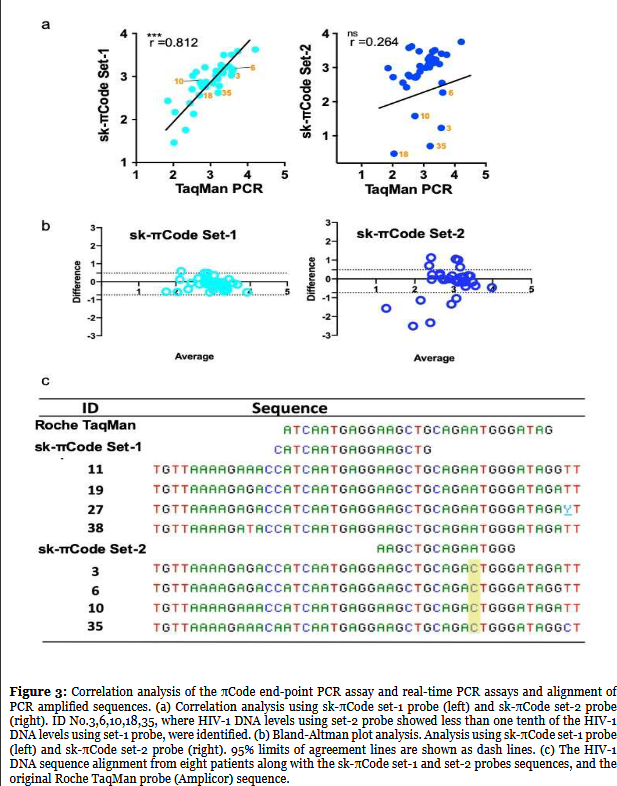
A correlation analysis was performed between πCode end-point PCR assay and real-time PCR assay using the data from Table 1. As expected, a strong correlation was confirmed between the πCode assay with sk-πCode set-1 probe and real-time PCR (r=0.81, 95%CI: 0.6596-0.9004, p<0.0001), shown in the left figure of Figure 3a. There was no correlation between the πCode assay with sk-πCode set-2 probe and real-time PCR (r=0.26, 95%CI: -0.0709-0.545, p: ns), right figure of Figure 3a. To do further comparisons, a Bland- Altman plot analysis was conducted (Figure 3b), showing, as predicted, a great agreement of the πCode PCR assay using πCode set-1 probe and the real-time PCR assay, with most of the plotted values within the two 95% limit lines (left figure of Figure 3b). The πCode set-2 probe assay and real-time PCR assay did not show good agreement (right figure of Figure 3b).
| ID | sk-πCode set-1 | sk-πCode set-2 | TaqMan Probe |
|---|---|---|---|
| 1 | 271 | 962 | 71 |
| 2 | 1467 | 1704 | 2139 |
| 3 | 1064 | 17 | 3674 |
| 4 | 1608 | 1707 | 2436 |
| 5 | 1442 | 1352 | 2023 |
| 6 | 1446 | 185 | 4102 |
| 7 | 52 | 243 | 0 |
| 8 | 3833 | 2367 | 5467 |
| 9 | 239 | 263 | 279 |
| 10 | 733 | 38 | 524 |
| 11 | 4238 | 5662 | 16169 |
| 12 | 3151 | 3211 | 2277 |
| 13 | 1625 | 3001 | 711 |
| 14 | 1834 | 2946 | 1945 |
| 15 | 503 | 612 | 321 |
| 16 | 557 | 570 | 615 |
| 17 | 1706 | 1265 | 3198 |
| 18 | 148 | 3 | 111 |
| 19 | 1322 | 355 | 3867 |
| 20 | 102 | 15 | 0 |
| 21 | 875 | 2054 | 1578 |
| 22 | 567 | 1108 | 750 |
| 23 | 134 | 550 | 350 |
| 24 | 141 | 501 | 0 |
| 25 | 1042 | 3787 | 334 |
| 26 | 364 | 520 | 503 |
| 27 | 3178 | 2554 | 3721 |
| 28 | 1048 | 1747 | 1397 |
| 29 | 1265 | 4161 | 409 |
| 30 | 1043 | 1019 | 1541 |
| 31 | 644 | 515 | 565 |
| 32 | 29 | 526 | 104 |
| 33 | 1193 | 1477 | 2189 |
| 34 | 1345 | 1213 | 1417 |
| 35 | 416 | 5 | 1610 |
| 36 | 692 | 978 | 1152 |
| 37 | 746 | 766 | 755 |
| 38 | 601 | 1957 | 1723 |
| 39 | 57 | 361 | 214 |
There are substantial differences between the πCode assay analysis data of sk-πCode set-1 probe and set-2 probe, which might be related to each probe position target in the HIV-1 sequence. As shown in Figure 1a, LNA-TaqMan probe and sk-πCode set-1 are designed to hybridize almost identical regions with only one base difference. The sk-πCode set-2 probe targets a region 12 bases down stream of that of the sk-πCode set-1 probe site. The five samples with different values between πCode set-1 probe and πCode set-2 probe, ID Nos. 3,6,10,18, and 35 are indicated in red color in Figure 3a. Similarly, the five samples HIV-1 DNA copy numbers detected with the sk-πCode set-2 probe were poorly correlated with the copy numbers by real-time PCR (Figure 3a).
Identification of HIV-1 subtypes
Next, a sequence analysis of the amplified HIV-1 DNAs obtained from patient ID No. 3,6,10, and 35 was performed. For comparison, the sequence of another four patients’ samples (ID No. 11,19,27,38) with a similar HIV-1 copy number for both sk-πCode set-1 probe and set-2 probe was also analyzed. The identification of the sequence for ID No. 18 was unsuccessful, which might likely be due to the variations in HIV-1 DNA integrated in this patient genome. The sequence alignment of the remaining four inconsistent patients, ID No. 3,6,10, and 35, is shown in Figure 3c. There is a one base point mutation across all four patients’ sequences, which was different from the sequences of ID No. 11,19,27,38. The HIV-1 sequence of the four inconsistent patients contained a “C” base (highlighted in yellow in Figure 3c) to the corresponding “A” base in the sk-πCode set- 2 probe sequence. The sk-πCode set-2 probe sequence was identical to the four patients (ID No.11,19,27,38) HIV-1 sequences used for comparison, similar to the sk- πCode set-1 probe. The sequence analysis obtained from the eight patients clearly explains the reduction of HIV-1 DNA copy numbers in the four inconsistent patients (ID No.3, 6, 10, 35). The one base mismatched sequence in the four patients’ sequences may be the reason for the poor detection generated by the sk-πCode set-2 probe.
Comparison of detection sensitivity in real-time PCR and the πCode end-point PCR
A further sensitivity comparison of both assays was conducted, following a typical procedure for checking PCR detection sensitivity [64]. HIV-1 DNA extractions from three clinical samples IDs #40, #41, #42, were threefold serially diluted for this comparison. The πCode endpoint PCR analysis with 35 PCR cycles using sk-πCode set-1 probe is shown in Table 2. The πCode assay was able to detect up to 9-fold dilution for the three samples tested. Real-time PCR analysis with 50 PCR cycles is also shown in Table 2. The undiluted samples were detected by real-time PCR, but none of the diluted samples were detected. This comparison revealed that the πCode end-point PCR assay shows 9 times higher sensitivity than real-time PCR. After 50 PCR cycles with the πCode end-point PCR assay, HIV-1 DNA quantification using the standard curve was not possible due to the MFI signal generated from the samples reaching maximum detection level. However, the presence of HIV-1 in a qualitative analysis could be done. The data revealed that the πCode end-point PCR assay is 81 (34) times more sensitive than real-time PCR for sample #40, and 27 (33) times more sensitive than real-time PCR for ID#41 and ID#42 (Supplementary Table 2).
| ID | 3 times dilution series | sk-πCode set-1 (35 PCR cycles) |
TaqMan probe (50 PCR cycles) |
|---|---|---|---|
| HIV-1 DNA copy | HIV-1 DNA copy | ||
| No.40 | 1/1 | 3.7 | 9.3 |
| 1/3 | 1.28 | ND | |
| 1/9 | 0.45 | ND | |
| 1/27 | ND | ND | |
| No.41 | 1/1 | 1.66 | 7.84 |
| 1/3 | 1.34 | ND | |
| 1/9 | 0.57 | ND | |
| 1/27 | ND | ND | |
| No.42 | 1/1 | 3.96 | 9.54 |
| 1/3 | 1.43 | ND | |
| 1/9 | 0.55 | ND | |
| 1/27 | ND | ND |
Reproducibility of the πCode end-point PCR
After finding exceptionally superior sensitivity in the πCode end-point PCR, the reproducibility of this assay was evaluated (Table 3). Five clinical samples containing different amounts of HIV-1 DNA were tested. The HIV- 1 DNA copy number in these samples was detected independently three times and mean and standard deviation (SD) was calculated from these values. CV value was determined for each sample, and as shown on Table 3, they are all under 20%.
| ID | sk-piCode set-1 | |||||
|---|---|---|---|---|---|---|
| HIV-1 copy number in 1x106 cells | M (Mean) | SD (Standard Deviation) |
CV (Coefficient of Variation) |
|||
| 1 | 217 | 288 | 309 | 271 | 48.1 | 0.177 |
| 7 | 42 | 55 | 58 | 52 | 8.5 | 0.163 |
| 8 | 3378 | 4262 | 3860 | 3833 | 442.6 | 0.115 |
| 11 | 4790 | 4446 | 3478 | 4238 | 680.3 | 0.161 |
| 13 | 1456 | 1480 | 1939 | 1625 | 272.2 | 0.168 |
Detection specificity in real-time PCR assay and πCode end-point PCR assays
Lastly, specificity of each assay was evaluated (Table 4). In general, highly sensitive assays may show less specificity in the developed assay. As expected, both realtime and πCode end-point assays did not show any nonspecific detection in the HIV-2 sample or controls, since the primer set of sk145 and skcc1B is well known to not have any non-specific amplification in HIV-1 DNA and negative controls.
| ID | HIV infection | πCode End-Point PCR | ||
|---|---|---|---|---|
| Real Time | sk set-1 | sk set-2 | ||
| 291 | HIV-1 | 338 | 469 | 568 |
| 391 | HIV-2 | 0 | 0 | 0 |
| A | Negative Control | 0 | 0 | 0 |
| B | Negative Control | 0 | 0 | 0 |
| C | Negative Control | 0 | 0 | 0 |
| D | Negative Control | 0 | 0 | 0 |
| E | Negative Control | 0 | 0 | 0 |
Discussion
The future of HIV therapy aims to eradicate the latent reservoir of HIV DNA-infected cells in the body that remains despite current fully suppressive ART. Studies of such therapeutic strategies will require assays that are much more sensitive than the currently available techniques. In this report, we compared a newly developed end-point PCR assay platform (πCode assay) with a real-time PCR assay platform for detection of PCR amplified HIV-1 DNA.
In our HIV diagnostic laboratory, we occasionally face difficulty with HIV-1 DNA detection, especially when we receive samples from HIV-1 positive expectant mothers, because those mothers are often under intensive ART. We sometimes face negative test results in HIV-1 DNA even with the current advanced diagnostic method, GeneXpert (Cepheid), providing qualitative analysis data. The situation is more challenging when we receive the newborn babies’ samples from those positive mothers. When we have HIV-1 negative status in the babies’ samples, we have less confidence of the negative status of these newborn babies. It could be false negative as we were unable to detect the presence of HIV-1 DNA in the mothers’ peripheral blood samples [65,66].
In general, within 3-8 months after starting ART, particularly with regimes including integrase inhibitors, the plasma VL of most of the infected patients is less than the detection limit [19-21,23]. After reaching this status (suppressed levels of plasma VL), most patients plasma VL remain suppressed for long periods of time in most of the developed countries since most of the patients remain adherent to treatment. Therefore, plasma VL analysis is no longer useful to fully reflect optimal efficacy of ART for patient management. Instead, HIV-1 DNA levels have shown to be a better marker for efficacy of ART [6,11,22-31], but we need a more sensitive assay platform than the current gold standard real-time PCR assay for quantitative HIV-1 DNA detection.
Currently, most clinical and research laboratories use real-time PCR detection systems for any type of DNA and RNA quantification. Real-time PCR is recognized as the gold standard method for quantification of any nucleic acid obtained from any type of specimen, since it is a rapid, reliable, and reproducible method, with more than 6-log quantification dynamic range. There are numerous studies describing the advantages of realtime PCR systems, based on solid theoretical background [67-70]. However, several reports indicate some caution in analyzing the results from quantitative real-time PCR and conventional end-point PCR [71]. The reports make special reference to the detection sensitivity associated with real-time PCR assay platform [36-40]. Before the development of real-time PCR, end-point PCR detection platform had shown to have quantitative capability based on non-radioisotope detection methods, where a biotin labeled probe to capture PCR product and a chemiluminescent substrate for amplification of detection signal were used [72-75]. These publications (reported more than 25 years ago) demonstrated similar sensitivities to current real-time PCR assay. The End- Point PCR system is now recognized as an assay format for “enzyme-linked immunosorbent assay (ELISA)-PCR”. After refining and optimizing this system, the ELISA-PCR showed superior sensitivity to real-time PCR assay [36-40]. More recently, ddPCR method has been developed to achieve superior sensitivity with quantification capability. The ddPCR is also based on end-point PCR platform [9,28,42,43,76]. There is a drawback associated with end-point PCR assay formats. The end-point PCR assay may have a limited dynamic range (not greater than 3-logs). However, HIV-1 DNA detection levels in patients under prolonged ART with totally suppressed plasma VL (HIV-1RNA<20 copies/mL), are consistently detected within the 3-log dynamic range, as reported by others [6,11,13,77]. Therefore, achieving a wide dynamic range is not essential for a HIV-1 DNA assay in most wellsuppressed ART patients. Achieving HIV-1 DNA detection method with high sensitivity is the most essential and urgent issue for HIV clinical diagnostic field.
Although there are some concerns about the sk145 and skcc1b primer set having less detection capability in non-subtype-B HIV-1 [45,78-80,], this primer set was the first FDA approved HIV-1 molecular diagnostic kit (Roche Amplicor HIV-1 detection assay) and has been used in every HIV-1 diagnostic lab in the world. Our lab is based in Sydney (Australia) and the great majority of patients have a HIV-1 subtype-B infection. Moreover, currently no quantitative real-time PCR assay is available for diagnostic laboratories, we used our quantitative real-time PCR assay with TaqMan probe based on sk145- skcc1b primer, as a gold standard of real-time PCR assay [47-63,81]. Therefore, in this research, we compared both assay formats, “πCode end-point PCR assay” and “real-time PCR assay” using the sk145 and skcc1b primer set.
After analysis of 39 HIV-1 infected patients, whose plasma viral load was less than 20 copies/mL for more than 2 years, we identified a strong correlation between the πCode end-point PCR assay using sk-πCode set-1 probe and the real-time PCR assay (r=0.81), since the sequence of the set-1 probe is almost identical to the LNA-TaqMan probe in our real-time PCR assay. The difference in the sequence of both probes was just one base. Our data may also suggest a possible reason why the original Roche Amplicor HIV-1 detection assay showed some difficulty to detect non-subtype-B HIV-1, after evaluation of the data obtained from the sk-πCode set-2. The sk-πCode set-2 targets HIV-1 DNA 12 bases downstream from the sk-πCode set-1. The original Roche TaqMan probe contains the same sequence of the sk- πCode set-2 probe. Our sequence alignment analysis of four inconsistent samples, which showed>10-fold less HIV-1 DNA copy number using sk-πCode set-2 probe compared with those values using the sk-πCode set- 1 probe, revealed that the amplified HIV-1 sequences contained one mismatched base at the same position across the four samples. This mismatched base might attribute the reduction in HIV-1 DNA copy numbers in the πCode end-point PCR assay. Furthermore, a subtype analysis for those patients identified only one sample out of the four was a non-subtype-B HIV-1. The other three samples were subtype-B HIV-1. Therefore, the data suggests that the original Roche Amplicor HIV- 1 detection assay might have some difficulty with even subtype-B HIV-1 sample. Roche Diagnostics used to provide an HIV-1 DNA confirmation test, which was used in almost all HIV Diagnostic and Research laboratories in the world for many years. However, later, there were reports which indicated that the primer set used in the test had issues to detect HIV-1 DNA in some clinical samples, especially from non-B subtype HIV-1 infections. The diagnostic kit was discontinued about 7 years ago. Our data suggests that one of the major reasons for the difficulty of the previous Roche kit to detect HIV-1 in certain patients could be related to the hybridization probe in the original kit. Our data also suggest that Roche long hybridization probe may have had some difficulty to detect even subtype-B HIV-1. This finding agrees with several previous reports identifying some difficulties to detect HIV-1 DNA in some subtype B samples using Roche Amplicor HIV-1 DNA PCR kit [45,46,78,79].
We also found that the TaqMan probe real-time PCR assay could not detect HIV-1 DNA in three samples, ID No. 7, 20, and 24. These three samples contained very low levels of HIV-1 DNA, while the πCode end-point PCR assay using the set-1 probe was able to detect the presence of HIV-1 DNA in peripheral whole blood samples of patients successfully treated with ART for more than two years, which suggests that the πCode endpoint PCR assay has superior sensitivity than real-time PCR. Further analysis using three-fold serially diluted clinical samples ID No. 40, 41, 42 in the πCode endpoint PCR assay with 50 cycles, which is equivalent to the real-time PCR analysis, showed at least >27-fold higher detection sensitivity than real-time PCR. Additionally, the πCode end-point PCR assay was very specific to HIV- 1 DNA detection, without any non-specific detection of HIV-2 DNA. We used an HIV-2 infected patient sample for this specificity evaluation, since HIV-1 and HIV-2 are from the same Lentivirus genus within the Retroviridae family [82]. The πCode end-point PCR assay was able to generate reproducible data with less than 20% CV. For the development of a clinical-type test based on PCR detection platform, if CV values are within 20% it indicates that the developed assay is stable with very little variation between each run [70,83]. This strongly confirms that the πCode end-point assay was able to generate reproducible data and is very stable. The πCode assay platform is based on the IntelliPlex1000 πCode Processor, an automated hybridizing and washing machine, and the PlexBio100 Analyzer for the fluorescent signal detection of the πCode MicroDiscs.
Conclusions
We demonstrated that HIV-1 DNA detection with the novel “πCode end-point PCR assay” is 100% sensitive, while the real-time PCR showed 92.3% in a head-to-head comparison in samples from HIV-1 infected individuals under ART, with suppressed levels of plasma VL for more than two years. We identified a strong correlation between the πCode end-point PCR assay using sk-πCode set-1 probe and the real-time PCR assay. TaqMan probe real-time PCR assay could not detect HIV-1 DNA in some samples with very low levels of HIV-1 DNA, while the πCode end-point PCR assay using the set-1 probe successfully detected the presence of HIV-1 DNA in all samples, confirming that the πCode end-point PCR assay has superior sensitivity than real-time PCR. Further analysis using three-fold serially diluted clinical samples in the πCode end-point PCR assay with 50 cycles, which is equivalent to the real-time PCR analysis, showed at least 27-fold higher detection sensitivity than real-time PCR. The πCode end-point PCR assay was very specific to HIV-1 DNA detection, without any non-specific detection of HIV-2 DNA, and generated reproducible data with less than 20% CV.
The πCode assay platform is based on the IntelliPlex1000 πCode Processor, an automated hybridizing and washing machine, and the PlexBio100 Analyzer for the fluorescent signal detection of the πCode MicroDiscs. Despite the high detection sensitivity achieved with the sk145 and skcc1b primer set, further optimization can accomplish a much higher sensitive HIV-1 DNA assay using different sets of primers, considering the limitation of the sk145 and skcc1b primer set to detect non-B clade HIV-1 subtypes [45,78-80]. In future studies, we will use a different primer set that can potentially achieve a successful detection of all HIV-1 subtypes plus maximum sensitivity for HIV-1 DNA detection. The current data, nonetheless, revealed that the πCode end-point PCR assay platform has great potential for developing highly specific and sensitive assays in various clinical diagnostic fields. The πCode end-point PCR assay format is a promising prospect for a new standard assay platform where ultrasensitive detection is required.
Author Contribution
Conceptualization, K.S., T.I., Y.L., and C.S.H; Methodology, K.S., T.I., Y.L., A.L., Y.L., S.M., and C.S.H.; Validation, K.S., T.I., Y.L., and C.S.H.; Formal Analysis, K.S., Z.L.; Investigation, K.S., T.I., Y.L., and C.S.H.; Rescues, K.S., J.Y., L.P.M., M.S. P.C.; Writing-Original Draft Preparation, K.S., A.L., T.I., Y.L., and C.S.H; Writing-Review & Editing, K.S., A.L., T.I., Y.L., J.Z., and C.S.H.
Acknowledgment
This research was funded by the St Vincent’s Clinic Foundation Research Grant and AMR Translational Research Grant.
Conflicts of Interest
The authors declare no conflict of interest.
References
2. Reda AA, Biadgilign S. Determinants of Adherence to Antiretroviral Therapy among HIV-Infected Patients in Africa. AIDS research and treatment. 2012 2012:574656.
3. Chun TW, Moir S, Fauci AS. HIV reservoirs as obstacles and opportunities for an HIV cure. Nature immunology. 2015 Jun;16(6):584.
4. Rouzioux C, Richman D. How to best measure HIV reservoirs? Current Opinion. HIV AIDS. 2013 8(3):170-5.
5. Henrich TJ, Deeks SG, Pillai SK. Measuring the Size of the Latent Human Immunodeficiency Virus Reservoir: The Present and Future of Evaluating Eradication Strategies. Journal of Infectious Diseases. 2017 215(suppl_3):S134-S41.
6. Kiselinova M, De Spiegelaere W, Buzon MJ, Malatinkova E, Lichterfeld M, Vandekerckhove L. Integrated and Total HIV-1 DNA Predict Ex Vivo Viral Outgrowth. PLoS Pathology. 2016 12(3):e1005472.
7. Avettand-Fenoel V, Bayan T, Gardiennet E, Boufassa F, Lopez P, Lecuroux C, Noel N, Tremeaux P, Monceaux V, Autran B, Meyer L, Saez-Cirion A, Lambotte O, Rouzioux C, Group CACS. Dynamics in HIV-DNA levels over time in HIV controllers. Journal of the International AIDS Society. 2019 22(1):e25221.
8. Véronique AF, Marie-Laure C, Stéphane B, Marianne B, Corinne F, Kadidia T, Marie-Christine A, Josiane W, Christine R, French Pediatric Cohort Study ANRS— CO 01 Group. LTR real-time PCR for HIV-1 DNA quantitation in blood cells for early diagnosis in infants born to seropositive mothers treated in HAART area (ANRS CO 01). Journal of medical virology. 2009 Feb;81(2):217-23.
9. Pannus P, Adams P, Willems E, Heyndrickx L, Florence E, Rutsaert S, De Spiegelaere W, Vandekerckhove L, Seguin-Devaux C, Vanham G. In-vitro viral suppressive capacity correlates with immune checkpoint marker expression on peripheral CD8+ T cells in treated HIVpositive patients. AIDS. 2019 33(3):387-98.
10. Zazzi M, Romano L, Catucci M, De Milito A, Almi P, Gonnelli A, Rubino M, Valensin PE. Low human immunodeficiency virus type 1 (HIV-1) DNA burden as a major cause for failure to detect HIV-1 DNA in clinical specimens by PCR. Journal of Clinical Microbiology. 1995 33(1):205-8.
11. Carr JM, Cheney KM, Coolen C, Davis A, Shaw D, Ferguson W, Chang G, Higgins G, Burrell C, Li P. Development of methods for coordinate measurement of total cell-associated and integrated human immunodeficiency virus type 1 (HIV-1) DNA forms in routine clinical samples: levels are not associated with clinical parameters, but low levels of integrated HIV- 1 DNA may be prognostic for continued successful therapy. J Clinical Microbiology. 2007 45(4):1288-97.
12. Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV- 1, even in patients on effective combination therapy. Nature medicine. 1999 May;5(5):512.
13. Pinzone MR, VanBelzen DJ, Weissman S, Bertuccio MP, Cannon L, Venanzi-Rullo E, Migueles S, Jones RB, Mota T, Joseph SB, Groen K, Pasternak AO, Hwang WT, Sherman B, Vourekas A, Nunnari G, O’Doherty U. Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nature communications. 2019 10(1):728.
14. Shan L, Siliciano RF. From reactivation of latent HIV-1 to elimination of the latent reservoir: the presence of multiple barriers to viral eradication. Bioessays. 2013 35(6):544-52.
15. Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nature Medicine. 2003 9(6):727-8.
16. Dickover RE, Donovan RM, Goldstein E, Cohen SH, Bolton V, Huth RG, Liu GZ, Carlson JR. Decreases in unintegrated HIV DNA are associated with antiretroviral therapy in AIDS patients. Journal of acquired immune deficiency syndromes. 1992;5(1):31-6.
17. Ho DD, Moudgil T, Alam M. Quantitation of human immunodeficiency virus type 1 in the blood of infected persons. New England journal of medicine. 1989 Dec 14;321(24):1621-5.
18. Josefsson L, King MS, Makitalo B, Brännström J, Shao W, Maldarelli F, Kearney MF, Hu WS, Chen J, Gaines H, Mellors JW. Majority of CD4+ T cells from peripheral blood of HIV-1–infected individuals contain only one HIV DNA molecule. Proceedings of the National Academy of Sciences. 2011 Jul 5;108(27):11199-204.
19. Hill AL, Rosenbloom DIS, Nowak MA, Siliciano RF. Insight into treatment of HIV infection from viral dynamics models. Immunological reviews. 2018 285(1):9-25.
20. Murray JM, Emery S, Kelleher AD, Law M, Chen J, Hazuda DJ, Nguyen BY, Teppler H, Cooper DA. Antiretroviral therapy with the integrase inhibitor raltegravir alters decay kinetics of HIV, significantly reducing the second phase. AIDS. 2007 21(17):2315-21.
21. Sanche S, Mesplede T, Sheehan NL, Li J, Nekka F. Exploring an alternative explanation for the second phase of viral decay: Infection of short-lived cells in a drug-limited compartment during HAART. PLoS One. 2018 13(7):e0198090.
22. Brodin J, Zanini F, Thebo L, Lanz C, Bratt G, Neher RA, Albert J. Establishment and stability of the latent HIV-1 DNA reservoir. Elife. 2016 5.
23. Burgard M, Blanche S, Jasseron C, Descamps P, Allemon MC, Ciraru-Vigneron N, Floch C, Heller- Roussin B, Lachassinne E, Mazy F, Warszawski J. Performance of HIV-1 DNA or HIV-1 RNA tests for early diagnosis of perinatal HIV-1 infection during antiretroviral prophylaxis. The Journal of pediatrics. 2012 Jan 1;160(1):60-6.
24. Gibellini D, Vitone F, Schiavone P, Ponti C, La Placa M, Re MC. Quantitative detection of human immunodeficiency virus type 1 (HIV-1) proviral DNA in peripheral blood mononuclear cells by SYBR green real-time PCR technique. Journal of Clinical Virology. 2004 29(4):282-9.
25. Hong F, Jacobs JL, Aga E, Cillo AR, Fyne E, Koontz DL, Zheng L, Mellors JW. Associations between HIV- 1 DNA copy number, proviral transcriptional activity, and plasma viremia in individuals off or on suppressive antiretroviral therapy. Virology. 2018 521:51-7.
26. Malnati MS, Scarlatti G, Gatto F, Salvatori F, Cassina G, Rutigliano T, Volpi R, Lusso P. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nature Protocols. 2008 3(7):1240-8.
27. Mortier V, Demecheleer E, Staelens D, Schauvliege M, Dauwe K, Dinakis S, Hebberecht L, Vancoillie L, Verhofstede C. Quantification of total HIV-1 DNA in buffy coat cells, feasibility and potential added value for clinical follow-up of HIV-1 infected patients on ART. Journal of Clinical Virology. 2018 106:58-63.
28. Rouzioux C, Avettand-Fenoel V. Total HIV DNA: a global marker of HIV persistence. Retrovirology. 2018 15(1):30.
29. Rouzioux C, Tremeaux P, Avettand-Fenoel V. HIV DNA: a clinical marker of HIV reservoirs. Current Opinion in HIV and AIDS. 2018 13(5):389-94.
30. Thomas J, Ruggiero A, Procopio FA, Pantaleo G, Paxton WA, Pollakis G. Comparative analysis and generation of a robust HIV-1 DNA quantification assay. Journal of Virological Methods. 2019 263:24-31.
31. van der Sluis RM, van Montfort T, Centlivre M, Schopman NC, Cornelissen M, Sanders RW, Berkhout B, Jeeninga RE, Paxton WA, Pollakis G. Quantitation of HIV-1 DNA with a sensitive TaqMan assay that has broad subtype specificity. Journal of Virological Methods. 2013 187(1):94-102.
32. Ceffa S, Luhanga R, Andreotti M, Brambilla D, Erba F, Jere H, Mancinelli S, Giuliano M, Palombi L, Marazzi MC. Comparison of the Cepheid GeneXpert and Abbott M2000 HIV-1 real time molecular assays for monitoring HIV-1 viral load and detecting HIV-1 infection. Journal of Virology Methods. 2016 229:35-9.
33. Mauk M, Song J, Bau HH, Gross R, Bushman FD, Collman RG, Liu C. Miniaturized devices for point of care molecular detection of HIV. Lab on a chip. 2017 17(3):382-94.
34. Nash M, Huddart S, Badar S, Baliga S, Saravu K, Pai M. Performance of the Xpert HIV-1 Viral Load Assay: a Systematic Review and Meta-analysis. Journal of Clinical Microbiology. 2018 56(4).
35. Bastien P, Procop GW, Reischl U. Quantitative realtime PCR is not more sensitive than “conventional” PCR. Journal of Clinical Microbiology. 2008 Jun 1;46(6):1897-900.
36. Mallet F, Hebrard C, Brand D, Chapuis E, Cros P, Allibert P, Besnier JM, Barin F, Mandrand B. Enzymelinked oligosorbent assay for detection of polymerase chain reaction-amplified human immunodeficiency virus type 1. Journal of Clinical Microbiology. 1993 31(6):1444-9.
37. Alborzi A, Pourabbas B, Shahian F, Mardaneh J, Pouladfar GR, Ziyaeyan M. Detection of Leishmania infantum kinetoplast DNA in the whole blood of asymptomatic individuals by PCR-ELISA and comparison with other infection markers in endemic areas, southern Iran. The American journal of tropical medicine and hygiene. 2008 Dec 1;79(6):839-42.
38. De Doncker S, Hutse V, Abdellati S, Rijal S, Singh Karki BM, Decuypere S, Jacquet D, Le Ray D, Boelaert M, Koirala S, Dujardin JC. A new PCR—ELISA for diagnosis of visceral leishmaniasis in blood of HIVnegative subjects. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2005 Jan 1;99(1):25- 31.
39. Li H, Dummer JS, Estes WR, Meng S, Wright PF, Tang YW. Measurement of human cytomegalovirus loads by quantitative real-time PCR for monitoring clinical intervention in transplant recipients. Journal of Clinical Microbiology. 2003 41(1):187-91.
40. Medeiros FA, Gomes LI, Oliveira E, de Souza CS, Mourao MV, Cota GF, Marques LH, Carneiro M, Rabello A. Development and Validation of a PCR-ELISA for the Diagnosis of Symptomatic and Asymptomatic Infection by Leishmania (Leishmania) infantum. Journal of tropical medicine. 2017 2017:7364854.
41. Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013 8(4):e55943.
42. Bosman KJ, Wensing AM, Pijning AE, van Snippenberg WJ, van Ham PM, de Jong DM, Hoepelman AI, Nijhuis M. Development of sensitive ddPCR assays to reliably quantify the proviral DNA reservoir in all common circulating HIV subtypes and recombinant forms. Journal of the International AIDS Society. 2018 21(9):e25185.
43. Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, Bertagnolli LN, Capoferri AA, Kufera JT, Timmons A, Nobles C, Gregg J, Wada N, Ho YC, Zhang H, Margolick JB, Blankson JN, Deeks SG, Bushman FD, Siliciano JD, Laird GM, Siliciano RF. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature. 2019 566(7742):120-5.
44. Kao JH, Lin CY, Chuang WL, Cheng YY, Hu JY, Liang WK, Friebe P, Palmer S, Huang CS. Clinical evaluation of IntelliPlex(TM) HCV genotyping kit for hepatitis C virus genotyping. Diagnostic microbiology and infectious disease. 2019.
45. Germer JJ, Gerads TM, Mandrekar JN, Mitchell PS, Yao JD. Detection of HIV-1 proviral DNA with the AMPLICOR HIV-1 DNA Test, version 1.5, following sample processing by the MagNA Pure LC instrument. Journal of Clinical Virology. 2006 37(3):195-8.
46. Lyamuya E, Olausson-Hansson E, Albert J, Mhalu F, Biberfeld G. Evaluation of a prototype Amplicor PCR assay for detection of human immunodeficiency virus type 1 DNA in blood samples from Tanzanian adults infected with HIV-1 subtypes A, C and D. Journal of Clinical Virology. 2000 17(1):57-63.
47. Hey-Nguyen WJ, Xu Y, Pearson CF, Bailey M, Suzuki K, Tantau R, Obeid S, Milner B, Field A, Carr A, Bloch M, Cooper DA, Kelleher AD, Zaunders JJ, Koelsch KK. Quantification of Residual Germinal Center Activity and HIV-1 DNA and RNA Levels Using Fine Needle Biopsies of Lymph Nodes During Antiretroviral Therapy. AIDS Reseaerch Human Retroviruses. 2017.
48. Koelsch K, Boesecke C, McBride K, Gelgor L, Fahey P, Natarajan V, Baker D, Bloch M, Murray J, Zaunders J, Emery S, Cooper D, Kelleher A. Impact of treatment with raltegravir during primary or chronic HIV infection on RNA decay characteristics and the HIV viral reservoir. AIDS. 2011 25(17):2069-78.
49. Koelsch KK, Rasmussen TA, Hey-Nguyen WJ, Pearson C, Xu Y, Bailey M, Marks KH, Sasson SC, Taylor MS, Tantau R, Obeid S, Milner B, Morrissey O, Pinto AN, Suzuki K, Busch MP, Keating SM, Kaiser P, Yukl S, Wong JK, Hiener BM, Palmer S, Zaunders J, Post JJ, Chan DJ, Avery S, Milliken ST, Kelleher AD, Lewin SR, Cooper DA. Impact of Allogeneic Hematopoietic Stem Cell Transplantation on the HIV Reservoir and Immune Response in 3 HIV-Infected Individuals. Journal of Acquire Immune Deficiency Syndrome. 2017 75(3):328-37.
50. McBride K, Xu Y, Bailey M, Seddiki N, Suzuki K, Murray JM, Gao Y, Yan C, Cooper DA, Kelleher AD, Koelsch KK, Zaunders J. The majority of HIV type 1 DNA in circulating CD4+ T lymphocytes is present in non-gut-homing resting memory CD4+ T cells. AIDS Research Human Retroviruses. 2013 29(10):1330-9.
51. Murray DD, Suzuki K, Law M, Trebicka J, Neuhaus J, Wentworth D, Johnson M, Vjecha MJ, Kelleher AD, Emery S, Insight E, Groups SS. Circulating microRNAs in Sera Correlate with Soluble Biomarkers of Immune Activation but Do Not Predict Mortality in ART Treated Individuals with HIV-1 Infection: A Case Control Study. PLoS One. 2015 10(10):e0139981.
52. Murray J, Zaunders J, McBride K, Xu Y, Bailey M, Suzuki K, Cooper D, Emery S, Kelleher A, Koelsch K. HIV DNA Subspecies Persist in both Activated and Resting Memory CD4+ T Cells during Antiretroviral Therapy. Journal of Virology. 2014 88(6):3516-26.
53. Murray JM, McBride K, Boesecke C, Bailey M, Amin J, Suzuki K, Baker D, Zaunders JJ, Emery S, Cooper DA, Koelsch KK, Kelleher AD, Pint Study T. Integrated HIV DNA accumulates prior to treatment while episomal HIV DNA records ongoing transmission afterwards. AIDS. 2012 26(5):543-50.
54. Zaunders J, Dyer WB, Churchill M, Munier CM, Cunningham PH, Suzuki K, McBride K, Hey-Nguyen W, Koelsch K, Wang B, Hiener B. Possible clearance of transfusion-acquired nef/LTR-deleted attenuated HIV-1 infection by an elite controller with CCR5 Δ32 heterozygous and HLA-B57 genotype. Journal of virus eradication. 2019 Apr;5(2):73..
55. Zaunders JJ, Ip S, Munier ML, Kaufmann DE, Suzuki K, Brereton C, Sasson SC, Seddiki N, Koelsch K, Landay A, Grey P, Finlayson R, Kaldor J, Rosenberg ES, Walker BD, Fazekas de St Groth B, Cooper DA, Kelleher AD. Infection of CD127+ (interleukin-7 receptor+) CD4+ cells and overexpression of CTLA-4 are linked to loss of antigen-specific CD4 T cells during primary human immunodeficiency virus type 1 infection. Journal of Virology. 2006 80(20):10162-72.
56. Higaki K, Hirao M, Kawana-Tachikawa A, Iriguchi S, Kumagai A, Ueda N, Bo W, Kamibayashi S, Watanabe A, Nakauchi H, Suzuki K. Generation of HIV-resistant macrophages from IPSCs by using transcriptional gene silencing and promoter-targeted RNA. Molecular Therapy-Nucleic Acids. 2018 Sep 7;12:793-804.
57. Murray DD, Suzuki K, Law M, Trebicka J, Neuhaus Nordwall J, Johnson M, Vjecha MJ, Kelleher AD, Emery S. Circulating miR-122 and miR-200a as biomarkers for fatal liver disease in ART-treated, HIV-1-infected individuals. Scientific Reports. 2017 7(1):10934.
58. Suzuki K, Ahlenstiel C, Marks K, Kelleher AD. Promoter Targeting RNAs: Unexpected Contributors to the Control of HIV-1 Transcription. Molecular Therapy Nucleic Acids. 2015 4:e222.
59. Suzuki K, Hattori S, Marks K, Ahlenstiel C, Maeda Y, Ishida T, Millington M, Boyd M, Symonds G, Cooper D, Okada S, Kelleher A. Promoter Targeting shRNA Suppresses HIV-1 Infection In vivo Through Transcriptional Gene Silencing. Molecular Therapy Nucleic Acids. 2013 2:e137 [E pub - December].
60. Suzuki K, Ishida T, Yamagishi M, Ahlenstiel C, Swaminathan S, Marks K, Murray D, McCartney E, Beard M, Alexander M, Purcell D, Cooper D, Watanabe T, Kelleher A. Transcriptional gene silencing of HIV-1 through promoter targeted RNA is highly specific. RNA Biology. 2011 8(6):1035-46.
61. Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper D, Rao S, Kelleher A. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. Journal of Biological Chemistry. 2008 283 23353-63.
62. Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, Kelleher A. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. Journal of RNAi and Gene Silencing. 2005 1(2):66-78.
63. Yamagishi M, Ishida T, Miyake A, Cooper D, Kelleher A, Suzuki K, Watanabe T. Retroviral delivery of promoter-targeted shRNA induces long-term silencing of HIV-1 transcription. Microbes Infection. 2009 11(4):500-8.
64. Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992 13(3):444-9.
65. Mazanderani AH, du Plessis NM, Thomas WN, Venter E, Avenant T. Loss of detectability and indeterminate results: Challenges facing HIV infant diagnosis in South Africa’s expanding ART programme. South African Medical Journal. 2014;104(8):574-7.
66. WHO. WHO Recommendations on the Diagnosis of HIV Infection in Infants and Children. WHO Guidelines Approved by the Guidelines Review Committee. Geneva 2010.
67. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry. 2009 55(4):611-22.
68. Dooms M, Chango A, Abdel-Nour A. Quantitative PCR (qPCR) and the guide to good practices MIQE: adapting and relevance in the clinical biology context. InAnnales de biologie clinique 2014 (Vol. 72, No. 3, pp. 265-269).
69. Raymaekers M, Smets R, Maes B, Cartuyvels R. Checklist for optimization and validation of real-time PCR assays. Journal of Clinical Laboratory Analysis. 2009 23(3):145-51.
70. Tichopad A, Dilger M, Schwarz G, Pfaffl MW. Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Research. 2003 31(20):e122.
71. Apfalter P, Reischl U, Hammerschlag MR. In-house nucleic acid amplification assays in research: how much quality control is needed before one can rely upon the results. Journal of clinical microbiology. 2005 Dec 1;43(12):5835-41.
72. Suzuki K, Craddock B, Kano T, Steigbigel R. Colorimetric reverse transcriptase assay for HIV-1. Journal of Virology Methods. 1993 41(1):21-8.
73. Suzuki K, Craddock BP, Okamoto N, Kano T, Steigbigel RT. Detection of Human Immunodeficiency Virus (HIV) by Colorimetric Assay for Reverse Transcriptase Activity on Magnetic Beads. Biotechnology & Applied Biochemistry. 1993 18(Part 1):37-44.
74. Suzuki K, Okamoto N, Kano T. Colorimetric detection for PCR amplified HIV-1 DNA using magnetic beads. Journal of Virological Methods. 1993 41(3):341-50..
75. Suzuki K, Okamoto N, Watanabe S, Kano T. Chemiluminescent microtiter method for detecting PCR amplified HIV-1 DNA. Journal of Virological Methods. 1992 38(1):113-22.
76. Bachmann N, von Siebenthal C, Vongrad V, Turk T, Neumann K, Beerenwinkel N, Bogojeska J, Fellay J, Roth V, Kok YL, Thorball CW, Borghesi A, Parbhoo S, Wieser M, Boni J, Perreau M, Klimkait T, Yerly S, Battegay M, Rauch A, Hoffmann M, Bernasconi E, Cavassini M, Kouyos RD, Gunthard HF, Metzner KJ, Swiss HIVCS. Determinants of HIV-1 reservoir size and long-term dynamics during suppressive ART. Nature communications. 2019 10(1):3193.
77. Chaillon A, Gianella S, Lada SM, Perez-Santiago J, Jordan P, Ignacio C, Karris M, Richman DD, Mehta SR, Little SJ, Wertheim JO. Size, composition, and evolution of HIV DNA populations during early antiretroviral therapy and intensification with maraviroc. Journal of virology. 2018 Feb 1;92(3):e01589-17.
78. Bøgh M, Machuca R, Gerstoft J, Pedersen C, Obel N, Kvinesdal B, Nielsen H, Nielsen C. Subtype-specific problems with qualitative Amplicor HIV-1 DNA PCR test. Journal of clinical virology. 2001 Feb 1;20(3):149-53.
79. Cunningham P, Marriott D, Harris C, Hancock S, Carr A, Cooper D. False negative HIV-1 proviral DNA polymerase chain reaction in a patient with primary infection acquired in Thailand. Journal of Clinical Virology. 2003 26(2):163-9.
80. Jackson JB, Piwowar EM, Parsons J, Kataaha P, Bihibwa G, Onecan J, Kabengera S, Kennedy SD, Butcher A. Detection of human immunodeficiency virus type 1 (HIV-1) DNA and RNA sequences in HIV-1 antibody-positive blood donors in Uganda by the Roche AMPLICOR assay. Journal of Clinical Microbiology. 1997 35(4):873-6.
81. Hey-Nguyen WJ, Bailey M, Xu Y, Suzuki K, Van Bockel D, Finlayson R, Leigh Brown A, Carr A, Cooper DA, Kelleher AD, Koelsch KK, Zaunders JJ. HIV-1 DNA Is Maintained in Antigen-Specific CD4+ T Cell Subsets in Patients on Long-Term Antiretroviral Therapy Regardless of Recurrent Antigen Exposure. AIDS Reseaerch Human Retroviruses. 2019 35(1):112-20.
82. German Advisory Committee Blood SAoPTbB. Human Immunodeficiency Virus (HIV). Transfusion Medical Hemotherapay. 2016 43(3):203-22.
83. Constantine NT, Callahan J, Watts DM. Retroviral Testing: essential for Quality Control and Laboratory Diagnosis: CRC Press, Inc.; 1992.