Abstract
Background: The dissemination of the extended spectrum β-lactamases (ESBL) producing E. coli poses a significant public health problem. Understanding the efficiency and frequency of horizontal gene transfer via conjugation of ESBL producing E. coli is imperative towards devising prevention and control measures. This study compared the frequencies and efficiencies of horizontal blaCTX-M gene transfer via conjugation among Escherichia coli isolates from urine and gastrointestinal tract (GIT) of patients with urinary tract infection (UTI), their animals and environment.
Methods: Horizontal blaCTX-M gene transfer via conjugation by a broth mating experiment was performed using 50 confirmed ESBL producing E. coli isolates as donors and Escherichia coli J53 (F−, met, pro, Azr), as the recipient. The transconjugants were detected and their frequencies and efficiencies of conjugation were measured and compared between ESBL producing E. coli isolates multi-sourced from urine, GIT, animals and
environment. Antimicrobial susceptibility testing of all resulting transconjugants was performed. DNA was extracted from all transconjugants to confirm the presence and the acquisition of blaCTX-M gene.
Results: Out of 50 ESBL producing E. coli isolates harboring blaCTX-M gene, 37 (74.0%) successfully exercised horizontal gene transfer through conjugation. All transconjugants were confirmed phenotypically and genotypically by PCR. Of note, all of the isolates from environment 100.0% (7/7) performed conjugation, exhibiting the highest transfer efficiency, followed by isolates from urine and animals, with the conjugation
transfer efficiency of 77.8% (14/18) and 76.1% (10/13), respectively. The isolates from the environment conjugated with a significant more efficiency than those from the GIT [Two-sample test of proportions; p-value = 0.0119]. The overall conjugation transfer frequencies ranged from 0.4 × 10-14 – 5.5 × 10-11 per donor cells with the highest median conjugation transfer frequency observed among isolates from animal (3.23 × 10-12 [IQR: 0.70 × 10-12 – 7.22 × 10-12]) followed by that of isolates from the environment (1.60 × 10-12 [IQR: 0.30 × 10-12 – 5.0 × 10-12]).
Conclusion: ESBL producing E. coli from human, animals and environment exercises horizontal blaCTX-M gene transfer efficiently with the highest occurrence among isolates from the environment and animals. The antimicrobial resistance control and prevention strategies should be widened up to explore strategies to prevent horizontal AMR gene transfer.
Keywords
Horizontal gene transfer, Conjugation, ESBL –producing E. coli, blaCTX-M gene
Introduction
World Health Organization (WHO) considers the extended spectrum β-lactamase producing Enterobactericeae including E. coli as a critical priority for antibiotic development [1] and these pathogens are regarded as a serious threat by the US Centers for Disease Control [2]. The dissemination of extended spectrum β-lactamase (ESBL) producing E. coli poses a significant problem since it is the commonest causative agent of urinary tract infection (UTI) and is described to be the highest ESBL producing strain with blaCTX-M gene acquired via horizontal gene transfer [3,4]. The emergence of ESBL producing E. coli limits treatment options for UTI [5]. Furthermore, it has been reported that E. coli obtains antimicrobial resistance faster than other microorganisms [6].
Reports show that the ESBL genes such as blaCTX-M, blaTEM and blaSHV have disseminated worldwide [3], of which blaCTX-M has been reported to be mostly disseminated through horizontal gene transfer [3]. Several genetic mechanisms have been involved in the spread of antimicrobial resistance genes and virulence genes, but conjugation is thought to have the greatest influence [4]. The mobile genetic elements such as conjugative plasmid, intergrons, and transposon are involved in dissemination of the antimicrobial resistance genes as well as virulence genes [3]. However, as compared to transformation and transduction, conjugation has been reported to be the mostly associated horizontal gene transfer mechanism via conjugative plasmid as the main vehicle for dissemination of antimicrobial resistance and virulence genes in both hospital and community settings [3,4]. Furthermore, studies have reported the horizontal gene transfer via conjugation disseminating antimicrobial resistance genes as well as virulence genes in enterobacteriaceae to occur in the human gut [7], animals [8] and environments [9]. This facilitates the spread of ESBL-producing E. coli across various ecological niches all over the globe and necessitates the surveillance of antimicrobial resistance to human, animals and environments.
Understanding the frequency and efficiency of horizontal gene transfer via conjugation among ESBL producing E. coli is imperative towards generating a better understanding of the mechanisms of dissemination of antimicrobial resistance genes in bacteria, an insight that is useful towards devising effective monitoring and prevention strategies against the dissemination of antimicrobial resistance genes. There is a high prevalence of the ESBL producing E. coli causing UTI and among those colonizing the GIT of human, animals, and environments [3,4,10-13], suggesting the possibility of horizontal gene transfer for dissemination of the blaCTX-M gene via conjugation within these bacterial populations. However, there is scarce information regarding the frequency and efficiency of conjugation exercised by ESBL producing E. coli from urine, gastrointestinal tract, animals, and environments of patients with urinary tract infection. Therefore, this study aimed to determine and compare the frequencies and efficiencies of horizontal blaCTX-M gene transfer via conjugation among E. coli isolates from urine and gastrointestinal tract (GIT) of patients with UTI, their animals, and environment.
Isolates, Materials and Methods
Study isolates
A total of 50 ESBL producing E. coli were used as donors for horizontal gene transfer via conjugation with plasmid carrying blaCTX-M gene to the Escherichia coli J53 (F−, met, pro, Azr), a mutant strain of E. coli and a kind gift from the Institute of Medical Microbiology, Giessen, Germany, was used as a recipient [14]. Of the 50 ESBL producing E. coli isolates, 18 isolates were from urine of patients with UTI, 12 isolates were from GIT of patients with UTI, 13 were from the animals kept by these patients, and 7 isolates were from environment of these patients. All the 50 ESBL-producing E. coli donors were confirmed phenotypically and genotypically by PCR to harbor blaCTX-M gene. All the donors were activated overnight in Brain Heart infusion broth at 37°C ready for use in conjugation by a broth mating experiment.
Horizontal blaCTX-M gene transfer via conjugation experiment
Conjugation of blaCTX-M gene from donor to recipient was performed using broth mating experiment as previously described [15] with slight modification. Briefly, the recipient strain which lacks blaCTX-M gene was prepared by streaking Escherichia coli J53 in Muller Hinton Agar (Himedia, India) plates supplemented with 100 µg/mL NaN3 [Sodium Azide], while donor strains were selected on Muller Hinton agar plates supplemented with 2 µg/mL Cefotaxime. From these, fresh overnight donor and recipient strains were prepared by picking single colonies emulsified in 10 mL Brain Heart Infusion (BHI) broth using different eppendorf tubes and incubated for overnight at 37°C. After exactly 12 hours, equal volumes (500 µL each) of donor and recipient strains were immediately mixed in 1.5 mL eppendorf tubes previously labeled transconjugant (Tc) while 1000 µL of donor strain were added in fresh tubes of similar volume to be separately selected on Muller Hinton agar plates supplemented with 2 µg/mL Cefotaxime and Muller Hinton agar plates supplemented with 100 µg/mL NaN3 as respective controls. All tubes were incubated at 37°C for 15 minutes, vortexed briefly, centrifuged at 12,000×g for 2 minutes and the pellet re-suspended in fresh 1000 µL Brain Heart infusion broth. Finally, 100 µL of 10−1 to 10−4 transconjugant cultures were double selected on Muller Hinton Agar plates supplemented with 100 µg/mL NaN3 and 2 µg/mL Cefotaxime. Conjugation frequency (transconjugants per donor cells) was reported as transconjugants per donor cells, with the denominator obtained from an initial volume of 1000 µL. Conjugation frequency was calculated for serially diluted transconjugant and donor cells, then reported as transconjugants per donor cells with an initial volume of 1000 µL donor cells. Conjugation efficiency was measured in percent (%) and was obtained by taking the proportion of donor isolates that managed to transfer the blaCTX-M gene to recipient cells divided by the total number of donor isolates tested.
Confirmation of horizontal blaCTX-M gene transfer via conjugation
Antibiotic susceptibility testing: Susceptibility testing of all resulting transconjugants was performed by the disk diffusion method on Mueller Hinton agar as recommended by the Clinical and Laboratory Standard Institute [16]. Antibiotics tested were: Cefotaxime (CTX) [2 ug/mL], Ciprofloxacin (CIP) [5 µg], Gentamicin (CN) [30 µg], Trimethoprim-Sulfamethoxazole (SXT) [1.25/23.75 μg] and Tetracycline (TET) [30 µg] (Hi-media, India).
DNA extraction of transconjugants: The boil lysate technique was used to extract bacterial DNA from isolates and selected transconjugants as previously reported [17] with slight modifications. Briefly, two colonies of overnight grown bacteria were suspended in brain heart infusion broth and incubated for six hours. The mixture was vortexed and boiled at 100°C in water-bath for 15 minutes. Tubes were centrifuged at 12000 rpm for 10 minutes and the DNA resuspended in 10 µl of nuclease free water. The quality of DNA was assessed by gel electrophoresis using TAE buffer while quantity was measured by qubit (Thermo Scientific). A minimum of 50 ng/µl was used for PCR reaction. Approximately 100 µl of DNA was aliquoted for storage at -20°C for further PCR amplification for detection of blaCTX-M gene.
PCR amplification for detection of blaCTX-M gene among transconjugants: Following DNA extraction, Polymerase Chain Reaction (PCR) was performed on thermal cycler machine (BIO-RAD, T100™, Singapore) for all transconjugants samples to amplify blaCTX-M gene using specific primers (Table 1) as previously described [8]. Briefly, 2 µl of DNA samples were added into PCR tubes containing a readily reconstituted master-mix (New England Biolabs) to make a final volume of 25 µl reaction mixture. PCR conditions were: initial denaturation 95°C for 15 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 40 seconds, and elongation at 72°C for 2 minutes. A final extension step followed at 72°C for 10 minutes. Finally, PCR products were detected in 1.5% agarose gel (Invitrogen from Fisher Scientific, UK), 90 Amp, 100V, for 45 minutes using TAE buffer and visualized under UV light.
|
Gene targets |
Primer name |
Primer sequences |
Amplicon |
|---|---|---|---|
|
blaCTX-M |
blaCTX-M_U_F blaCTX-M_U_R |
5’-ATGTGCAGYACCAGTAARGTKATGGC-3’ 5’-TGGGTRAARTARGTSACCAGAAYCAGCGG-3’ |
593 bp |
Quality control
Klebsiella pneumoniae ATCC 700603, Escherichia coli ATCC 35218 and a clinical isolate of non ESBL Escherichia coli were used as control strains. These control strains were used to check the performance of media, antibiotic discs, as well as PCR experiments for amplification and detection of ESBL alleles.
Data management and statistical analysis
Data from isolates such as identification number, isolate name, source of isolation, susceptibility pattern, conjugative frequency, and conjugative efficiency were recorded in the log book and then entered into the computer using Microsoft excel. Data were imported to STATA software version 15 for analysis. Kruskal-Wallis equality-of-populations rank test was used to compare the conjugation frequencies of ESBL-producing E. coli donors to the recipient between various sources of these isolates. Two-sample test of proportions was performed to compare the conjugation efficiency (Proportion) between isolates from the GIT which is the habitat for commensals E. coli with those from urine, animals, and environment. In all analyses the significance level was set at a p-value less than 0.05.
Ethical clearance
The ethical clearance for this work was granted by the Joint Catholic University of Health and Allied Science –Bugando Medical Center (CUHAS-BMC) Ethical Review Committee (CREC).
Results
Conjugation efficiency and frequency of the blaCTX-M gene
Out of the 50 ESBL producing E. coli isolates harboring blaCTX-M gene, 37 (74.0%) successfully transferred the gene via horizontal gene transfer through conjugation (Conjugation efficiency). All transconjugants were positive phenotypically and genotypically for blaCTX-M gene by showing a band of 593 bp on PCR (Figure 1). Of note, isolates from environment, performed conjugation with the highest transfer efficiency, as all of the isolates, 100.0% (7/7) successfully exercised conjugation transfer of blaCTX-M gene, followed by the isolates from urine and animals, with the conjugation transfer efficiency of 77.8% (14/18) and 76.1% (10/13) respectively. Of note, isolates from the environment conjugated with a significant more efficiency than those from the GIT [Two-sample test of proportions; p-value = 0.0119] (Table 2). The overall conjugation transfer frequencies ranged from 0.4 × 10-14 – 5.5 × 10-11 per donor cells. The highest median conjugation transfer frequency was observed among isolates from animal (3.23 × 10-12 [IQR: 0.70 × 10-12 – 7.22 × 10-12]) followed by that of isolates from the environment (1.60×10-12 [IQR: 0.30 × 10-12 – 5.0 × 10-12]). There was no statistically significant difference between the conjugation frequencies observed among the ESBL producing E. coli multi-sourced from urine, GIT, animals, and environment [Kruskal-Wallis equality-of-populations rank test; p-value = 0.8116] (Table 3). This emphasizes that ESBL producing E. coli performs horizontal blaCTX-M gene via conjugation at comparable frequencies.
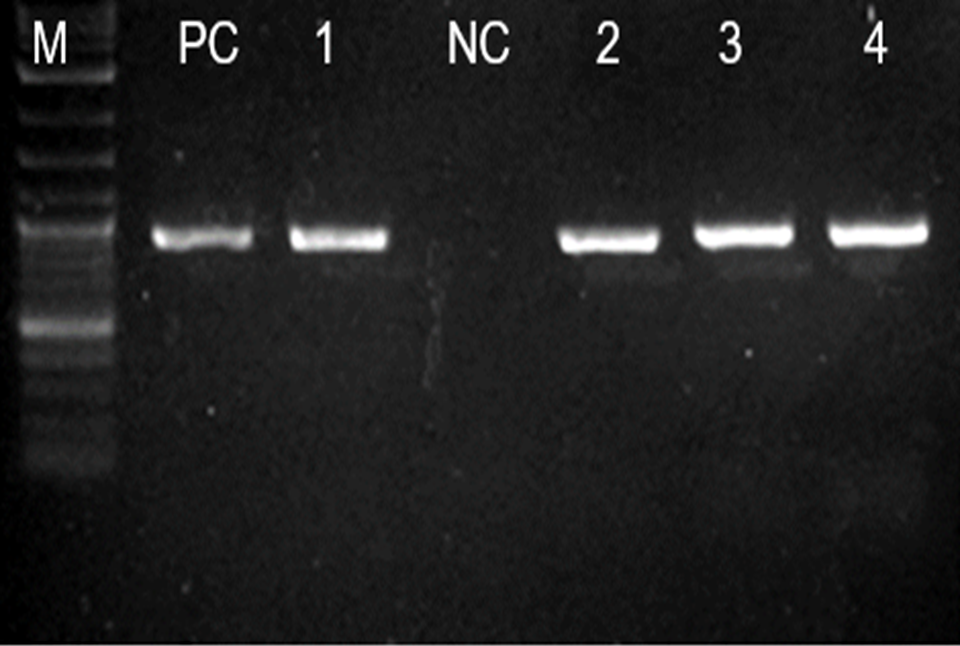
Figure 1. Gel image showing the 593 blaCTX-M gene PCR product in 4 transconjugants. Ladder: 100bp (New England BioLabs), Lane PC: Positive control blaCTX-M (K. pneumoniae ATCC 700603), Lane NC Negative control (Clinical isolate E. coli non ESBL), Lanes 1, 2, 3, and 4: blaCTX-M gene.
|
Source of ESBL – producing E. coli |
Transconjugants Detected |
p-value |
|
|
Yes |
No |
||
|
n (%) |
n (%) |
||
|
Overall |
37 (74.0) |
13 (26.0) |
- |
|
GIT |
6 (50.0) |
6 (50.0) |
Reference |
|
Animal |
10 (76.9) |
3 (23.1) |
0.0840 |
|
Urine |
14 (77.8) |
4 (22.2) |
0.0868 |
|
Environment |
7 (100.0) |
0 (0.0) |
0.0119 |
|
Source of ESBL – producing E. coli |
Median |
Interquartile Range [IQR] |
p-value |
|---|---|---|---|
|
Overall |
1.60 × 10-12 |
0.39 × 10-12 – 4.00 × 10-12 |
- |
|
GIT |
1.16 × 10-12 |
0.47 × 10-12 – 2.70 × 10-12 |
0.8116 |
|
Animal |
3.23 × 10-12 |
0.70 × 10-12 – 7.22 × 10-12 |
|
|
Urine |
0.91 × 10-12 |
0.30 × 10-12 – 5.50 × 10-12 |
|
|
Environment |
1.60 × 10-12 |
0.30 × 10-12 – 5.00 × 10-12 |
Antibiotic resistance phenotypes of donors from urine, GIT, animals, and environments and transconjugants
Of the 37 transconjugants detected, all (100.0%) expressed phenotypic drug resistance to Cefotaxime (CTX). Only 6 (16.2%) isolates expressed phenotypic resistance to Cefotaxime (CTX) alone yet their donor had also other antimicrobial resistance. Of note, there were 31 (83.8%) isolates that acquired and expressed antimicrobial resistance beyond Cefotaxime (CTX) from their donors. This includes resistance to ciprofloxacin (CIP), Gentamicin (CN), Trimethoprim- sulfamethoxazole (SXT), and Tetracycline (TET) in various combinations (Table 4).
|
SN |
Sample ID |
Sample Source |
Conjugation frequency |
Donors’ Resistance Phenotypes |
Transconjugants’ Resistance Phenotypes |
|
1 |
A1 |
ANIMAL |
0.37 × 10-12 |
CTX + CIP + CN |
CTX |
|
2 |
A2 |
ANIMAL |
0.39 × 10-12 |
CTX + CIP + CN |
CTX + CIP |
|
3 |
A3 |
ANIMAL |
8.00 × 10-12 |
CTX + CIP + CN |
CTX |
|
4 |
A4 |
ANIMAL |
0.07 × 10-11 |
CTX + CIP + CN |
CTX + CIP + CN |
|
5 |
A5 |
ANIMAL |
0.40 × 10-11 |
CTX + CIP + CN |
CTX + CIP + CN |
|
6 |
A6 |
ANIMAL |
3.86 × 10-12 |
CTX + CIP + CN |
CTX + CIP + CN |
|
7 |
A7 |
ANIMAL |
2.60 × 10-12 |
CTX + CIP + CN |
CTX + CIP + CN |
|
8 |
A8 |
ANIMAL |
0.86 × 10-12 |
CTX + CIP + CN |
CTX |
|
9 |
A9 |
ANIMAL |
6.44 × 10-12 |
CTX + CIP + CN |
CTX + CN |
|
10 |
A10 |
ANIMAL |
0.40 × 10-11 |
CTX + CIP + CN |
CTX + CN |
|
11 |
A11 |
ANIMAL |
- |
CTX + CIP + CN |
- |
|
12 |
A12 |
ANIMAL |
- |
CTX + CIP + CN |
- |
|
13 |
A13 |
ANIMAL |
- |
CTX + CIP + CN |
- |
|
14 |
G1 |
GIT |
0.47 × 10-12 |
CTX + SXT + CIP + CN + TET |
CTX + SXT + CIP + CN + TET |
|
15 |
G2 |
GIT |
1.71 × 10-12 |
CTX + CN |
CTX + CN |
|
16 |
G3 |
GIT |
1.45 ×10-11 |
CTX + SXT + TET |
CTX + TET |
|
17 |
G4 |
GIT |
0.60 × 10-12 |
CTX + SXT + CIP + CN + TET |
CTX + SXT + CIP + CN + TET |
|
18 |
G5 |
GIT |
0.27 × 10-11 |
CTX + SXT + TET |
CTX + SXT + TET |
|
19 |
G6 |
GIT |
0.20 × 10-12 |
CTX + SXT + CIP + CN |
CTX + CN |
|
20 |
G7 |
GIT |
- |
CTX + SXT + CIP + CN |
- |
|
21 |
G8 |
GIT |
- |
CTX + SXT + CIP |
- |
|
22 |
G9 |
GIT |
- |
CTX + SXT + CIP + CN |
- |
|
23 |
G10 |
GIT |
- |
CTX + SXT + TET |
- |
|
24 |
G11 |
GIT |
- |
CTX + CIP + CN |
- |
|
25 |
G12 |
GIT |
- |
CTX + SXT + CIP + CN |
- |
|
26 |
U1 |
URINE |
0.72 × 10-12 |
CTX + SXT + CIP + CN + TET |
CTX + SXT + CIP + CN + TET |
|
27 |
U2 |
URINE |
0.70 × 10-11 |
CTX + SXT + CIP + CN + TET |
CTX |
|
28 |
U3 |
URINE |
0.01 × 10-12 |
CTX + SXT + CN + TET |
CTX + SXT + CN + TET |
|
29 |
U4 |
URINE |
0.10 × 10-12 |
CTX + SXT + CN + TET + CIP |
CTX + TET + CIP |
|
30 |
U5 |
URINE |
0.11 × 10-11 |
CTX + SXT + CN + TET + CIP |
CTX + SXT + TET + CIP |
|
31 |
U6 |
URINE |
0.70 × 10-11 |
CTX + SXT + TET |
CTX |
|
32 |
U7 |
URINE |
0.10 × 10-12 |
CTX + SXT + CIP + TET |
CTX + CIP |
|
33 |
U8 |
URINE |
0.70 × 10-12 |
CTX + SXT + TET |
CTX + TET |
|
34 |
U9 |
URINE |
0.20 × 10-11 |
CTX + SXT |
CTX |
|
35 |
U10 |
URINE |
5.50 × 10-11 |
CTX + CIP + CN + TET |
CTX + CN |
|
36 |
U11 |
URINE |
0.30 × 10-12 |
CTX + SXT + TET |
CTX + TET |
|
37 |
U12 |
URINE |
0.30 × 10-11 |
CTX + SXT + TET |
CTX + TET |
|
38 |
U13 |
URINE |
3.45 × 10-13 |
CTX + SXT + CIP + CN |
CTX + CN |
|
39 |
U14 |
URINE |
0.50 × 10-12 |
CTX + SXT + CIP + TET |
CTX + CIP + TET |
|
40 |
U15 |
URINE |
- |
CTX + SXT + CIP + CN + TET |
- |
|
41 |
U16 |
URINE |
- |
CTX + SXT + CIP + CN |
- |
|
42 |
U17 |
URINE |
- |
CTX + SXT + CIP + TET |
- |
|
43 |
U18 |
URINE |
- |
CTX + SXT + CIP + TET |
- |
|
44 |
E1 |
ENVIRONMENT |
0.40 × 10-12 |
CTX + CIP + CN |
CTX + CIP + CN |
|
45 |
E2 |
ENVIRONMENT |
0.30 × 10-12 |
CTX + CIP + CN |
CTX + CIP + CN |
|
46 |
E3 |
ENVIRONMENT |
0.50 × 10-11 |
CTX + CIP + CN |
CTX + CIP + CN |
|
47 |
E4 |
ENVIRONMENT |
3.20 × 10-12 |
CTX + CIP + CN |
CTX + CIP + CN |
|
48 |
E5 |
ENVIRONMENT |
1.23 × 10-11 |
CTX + CIP + CN |
CTX + CIP + CN |
|
49 |
E6 |
ENVIRONMENT |
1.60 × 10-12 |
CTX + CIP + CN |
CTX + CIP + CN |
|
50 |
E7 |
ENVIRONMENT |
0.40 × 10-14 |
CTX + CIP + CN |
CTX + CIP + CN |
|
SXT: Trimethoprim-sulfamethoxazole; CIP: Ciprofloxacin; TE: Tetracycline; CN: Gentamicin; CTX: Cefotaxime |
|||||
Discussion
The findings from this study reveal that ESBL producing E. coli from urine and gastrointestinal tract of patients with UTI, their animals, and environment exercise horizontal blaCTX-M gene transfer efficiently with the highest occurrence among isolates from the environment and animals. Furthermore, these isolates showed comparable conjugation transfer frequencies. This emphasizes that conjugation attributes to horizontal gene transfer for propagation of ESBL genes for drug resistance and underpins the intensification of antimicrobial surveillance from human to animals to environments.
The findings show that 74.0% of the ESBL producing E. coli from urine and gastrointestinal tract of patients with UTI, animals kept by these patients and their environment performed horizontal gene transfer via conjugation, with a 100% efficiency for the isolates from environment. This finding is similar to studies done by Minja et al., and Pérez-Etayo et al., in which the 88.1% and 100% respectively of the tested ESBL-producing E. coli strains, were able to perform an efficient gene transfer [15,19]. Furthermore, in Minja et al., study all six ESBL producing E. coli isolates from the proper environment (soil) had a transfer efficiency of 100.0% similar to our finding. In our study the ESBL producing E. coli isolated from animals showed the highest conjugation transfer frequency. This highlights that environments and animals could be the most suitable ecological niche for ESBL-producing E. coli to mobilize and disseminate the antimicrobial resistance genes. Studies have shown that conjugation is one of the most important mechanisms for intra- and inter-species horizontal gene transfer, and it plays a significant role to accelerate the dispersal of antibiotic resistance genes [19-21]. Furthermore, ESBL genes have shown to be generally acquired by horizontal gene transfer [22] with some ESBL genes such as blaCTX-M gene mobilized from environmental bacteria [23]. For this reason, the fight against antibiotic resistance is a challenging and escalating problem liable to overwhelm our health systems. Several approaches are currently envisaged to combat the wide spread of antibiotic resistant pathogens particularly, by holding back the horizontal gene transfer. The continual search for specific conjugation inhibitors should be prioritized towards prevention of horizontal gene transfer of antimicrobial resistance as one of key target in the fight against the spread of antibiotic resistance genes [24].
When comparing the frequencies of conjugation for ESBL producing E. coli multi-sourced from urine, gastrointestinal tract of patients with UTI, their animals, and environment, we found there was no significant difference. This highlights that these ESBL producing E. coli perform horizontal blaCTX-M gene transfer via conjugation at comparable frequencies. This finding is also similar to the study done by Etayo et al., and Minja et al., which showed that the range of conjugation frequencies was nearly the same in all ESBL producing E. coli samples from various sources [15,18]. This habit fuels the acquisition of ESBL resistance genes across various ecological niches and gives the awaken call that the antimicrobial resistance surveillance, control and prevention strategies should be widen up to human, animals, and environment, as conjugation plays a major role in the spread of anti microbial resistance and the increasing occurrence of antibiotic resistance among pathogenic bacteria is considered a major problem for public health in recent decades [3,4,25-29].
In conclusion ESBL producing E. coli from urine and gastrointestinal tract of patients with UTI, their animals and environment exercise horizontal blaCTX-M gene transfer efficiently with the highest efficiency among isolates from the environment and highest frequency among isolates from animals. This emphasizes that conjugation appears to be a common phenomenon among ESBL producing E. coli and it attributes to horizontal gene transfer for propagation of ESBL genes for drug resistance. The antimicrobial resistance surveillance, control and prevention strategies should be widen up to human, animals and environment. Further studies are warranted in exploring the mobile genetic elements circulating in the environments and animals and the search for specific conjugation inhibitors should be prioritized towards prevention of horizontal gene transfer of antimicrobial resistance as one of key target in the fight against the spread of antibiotic resistance genes.
Data Availability
The raw data for our findings supporting the conclusion of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Authors’ Contributions
AAM, CAM, BRK, and SEM designed the study. AAM, CAM, and MFM carried out the experiments. BRK, CAM, and MFM analyzed the results. AAM, CAM, and BRK wrote the manuscript. WB, AS, MTGH, MFM, SEM, BRK, critically reviewed the manuscript. All authors read and approved the final version of the manuscript.
Funding
This study was funded by the Holistic Approach to Unravel Antibacterial Resistance in East Africa (HATUA) project funded by the National Institute for Health Research, Medical Research Council and the Department of Health and Social Care, Award (MR/S004785/1). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that there is no existing competing of interest.
References
2. Centers for Disease Control and Prevention (United States) Antibiotic/Antimicrobial Resistance. Biggest Threats and Data. 2019 [(accessed on 20 June 2022)]; Available online: https://www.cdc.gov/drugresistance/biggest- threats.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fdrugresistance%2Fbig gest_threats.html#groupb.
3. Norman A, Hansen LH, Sørensen SJ. Conjugative plasmids: vessels of the communal gene pool. Philosophical Transactions of the Royal Society B: Biological Sciences. 2009 Aug 12;364(1527):2275-89.
4. Von Wintersdorff CJ, Penders J, Van Niekerk JM, Mills ND, Majumder S, Van Alphen LB, Savelkoul PH, Wolffs PF. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Frontiers In Microbiology. 2016 Feb 19;7:173.
5. Hassuna NA, Khairalla AS, Farahat EM, Hammad AM, Abdel-Fattah M. Molecular characterization of Extended-spectrum β lactamase-producing E. coli recovered from community-acquired urinary tract infections in Upper Egypt. Scientific Reports. 2020 Feb 17;10(1):1-8.
6. Ben Yahia H, Ben Sallem R, Tayh G, Klibi N, Ben Amor I, Gharsa H, et al. Detection of CTX-M-15 harboring Escherichia coli isolated from wild birds in Tunisia. BMC microbiology. 2018 Dec;18:26.
7. Smith HW. Transfer of antibiotic resistance from animal and human strains of Escherichia coli to resident E. coli in the alimentary tract of man. The Lancet. 1969 Jun 14;293(7607):1174-6.
8. Cavaco LM, Abatih E, Aarestrup FM, Guardabassi L. Selection and persistence of CTX-M-producing Escherichia coli in the intestinal flora of pigs treated with amoxicillin, ceftiofur, or cefquinome. Antimicrobial Agents And Chemotherapy. 2008 Oct;52(10):3612-6.
9. Amos GC, Hawkey P, Gaze WH, Wellington E. Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. Journal of Antimicrobial Chemotherapy. 2014;69(7):1785-91.
10. Mshana SE, Imirzalioglu C, Hain T, Domann E, Lyamuya EF, Chakraborty T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clinical Microbiology and Infection. 2011 Aug 1;17(8):1279-82.
11. Mshana SE, Falgenhauer L, Mirambo MM, Mushi MF, Moremi N, Julius R, Seni J, Imirzalioglu C, Matee M, Chakraborty T. Predictors of bl a CTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infectious Diseases. 2016 Dec;16:187.
12. Seni, J., Falgenhauer, L., Simeo, N., Mirambo, M.M., Imirzalioglu, C., Matee, et al. Multiple ESBL-producing Escherichia coli sequence types carrying quinolone and aminoglycoside resistance genes circulating in companion and domestic farm animals in Mwanza, Tanzania, harbor commonly occurring plasmids. Frontiers in Microbiology. 2016;7:142.
13. Moremi N, Manda EV, Falgenhauer L, Ghosh H, Imirzalioglu C, Matee M, et al. Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza, Tanzania. Frontiers in microbiology. 2016 Dec 1;7:1862.
14. Jacoby GA, Han P. Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. Journal of Clinical Microbiology. 1996 Apr;34(4):908-11.
15. Minja CA, Shirima G, Mshana SE. Conjugative plasmids disseminating ctx-m-15 among human, animals and the environment in Mwanza Tanzania: a need to intensify one health approach. Antibiotics. 2021 Jul 9;10(7):836.
16. Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Standard M100: Wayne, PA, USA, 2018.
17. Mgaya FX, Matee MI, Muhairwa AP, Hoza AS. Occurrence of multidrug resistant Escherichia coli in raw meat and cloaca swabs in poultry processed in slaughter slabs in Dar es Salaam, Tanzania. Antibiotics. 2021 Mar 24;10(4):343.
18. Pérez-Etayo L, González D, Vitas AI. The aquatic ecosystem, a good environment for the horizontal transfer of antimicrobial resistance and virulence-associated factors among extended spectrum β-lactamases producing E. coli. Microorganisms. 2020 Apr 15;8(4):568.
19. Mazel D, Davies J. Antibiotic resistance in microbes. Cellular and Molecular Life Sciences CMLS. 1999 Nov;56:742-54.
20. Barlow M. What antimicrobial resistance has taught us about horizontal gene transfer. Horizontal Gene Transfer: Genomes in Flux. 2009:397-411.
21. Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. Network analyses structure genetic diversity in independent genetic worlds. Proceedings of the National Academy of Sciences. 2010 Jan 5;107(1):127-32.
22. Malloy AM, Campos JM. Extended-spectrum beta-lactamases: a brief clinical update. The Pediatric Infectious Disease Journal. 2011 Dec 1;30(12):1092-3.
23. Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K, Huijsdens X. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerging Infectious Diseases. 2011 Jul;17(7):1216-22.
24. Cabezón Navarro ME, Cruz Calahorra FD, Arechaga Iturregui IM. Conjugation Inhibitors and Their Potential Use to Prevent Dissemination of Antibiotic Resistance Genes in Bacteria. Front Microbiol. 2017;8:2329.
25. Zatyka M, Thomas CM. Control of genes for conjugative transfer of plasmids and other mobile elements. FEMS Microbiology Reviews. 1998 Feb 1;21(4):291-319.
26. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001 May 1;29(9):e45.
27. Sørensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. Studying plasmid horizontal transfer in situ: a critical review. Nature Reviews Microbiology. 2005 Sep 1;3(9):700-10.
28. Bennett PM. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. British journal of pharmacology. 2008 Mar;153(S1):S347-57.
29. Thomsen MC, Ahrenfeldt J, Cisneros JL, Jurtz V, Larsen MV, Hasman H, et al. A bacterial analysis platform: an integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PloS One. 2016 Jun 21;11(6):e0157718.
