Commentary
For decades, the scientific community tried hard to identify the gene biomarkers whose mutations or regulations cause (better say are associated with) specific forms of cancer. For instance, the September 17th 2019 release of the Genomic Data Commons Data Portal [1] includes 3,142,246 mutations detected in 22,872 genes sequenced from 37,075 cases of cancers localized in 67 primary sites. The hope was (and still is) to treat cancer by restoring the normal sequence or/and expression level of the biomarker(s). The biomarkers are selected from the most frequently and broadly altered genes in large cohorts of patients affected by the same form of cancer. Problem is that the frequent alteration indicates that the biomarkers are not in the high priorities of the cell to maintain their normal sequence or/and expression, and, as minor players, their restoration might not be of much consequence.
Despite the very rich literature and the ambitious (and very well-funded) Cancer Genome Atlas Program (TCGA [2]), there is not yet a comprehensive explanation of cancer development, nor a perfect therapeutic solution. Most cancers occur from nowhere, without being genetically inherited or directly caused by a steady deficient diet (affecting the microbiome), exposure to ionizing radiation or carcinogenic toxins, or bad habit (like smoking), although such risk factors increase the chances of the “bad luck” [3,4]. Tumors are heterogeneous, composed of regions with distinct characteristics, some of them malignant, some others preserving the normal features of the tissue. On top of these, with all similarities, each human is unique and has a unique lifeline, so, although a trained pathologist can recognize the cancer type, the tumors are not identical, nor develop identically or respond identically to treatment.
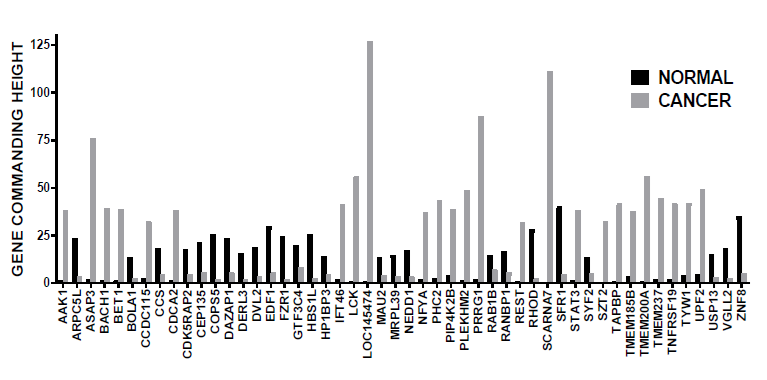
Therefore, instead of targeting the SAME alleged gene biomarker for all humans with a particular cancer form, we devised a method by which the cancer of the ACTUAL patient itself indicates what genes are NOW commanding it. We call these commanders “gene master regulators” (GMRs) and identify them by profiling the transcriptomes of tumor biopsies or blood samples (pending on the suspected cancer type) using RNA sequencing or microarray platforms. The method, consistent with our Genomic Fabric Paradigm [5], relies on original mathematical algorithm and software that establish the gene hierarchy based on their Gene Commanding Height (GCH). GCH is a composite measure of gene expression control and coordination with major functional pathways. The GMR tops the most controlled genes by the homeostatic mechanisms (because they are critical for the cell survival and phenotypic expression) whose expression regulates most functional pathways through coordination with the expression of many other genes. The GMR approach provides the most legitimate targets for cancer gene therapy. It is also personalized and time-sensitive because the GMR hierarchy is unique for each patient and changes slowly during cancer development.
In the commented [6] and other recent papers [7,8], we proved that cancer nuclei and surrounding normal tissue are governed by distinct GMRs (illustrated in Figure 1 for a case of prostate cancer). We have also tested on standard human cancer cell lines that manipulation of the expression of a gene has transcriptomic consequences consistent with its GCH score. As such, it is expected that targeting the GMRs of cancer nuclei from a tissue will selectively destroy the cancer cells with little consequences on the normal ones.
Acknowledgment
D.A.I. was supported by the Chancellor's Research Initiative (CRI) funding for the Center for Computational Systems Biology at the Prairie View A&M University".
References
2. https://www.cancer.gov/about-nci/organizationccg/research/structural-genomics/tcga
3. Tomasetti C, Li L, Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 2017;355(6331):1330-1334.
4. https://directorsblog.nih.gov/2017/04/04/randommutations-play-major-role-in-cancer
5. Iacobas DA. The Genomic Fabric Perspective on the Transcriptome between Universal Quantifiers and Personalized Genomic Medicine. Biological Theory 2016; 11(3): 123-137. DOI 10.1007/s13752-016-0245-3.
6. Iacobas S, Ede N, Iacobas DA. The Gene Master Regulators (GMR) Approach Provides Legitimate Targets for Personalized, Time-Sensitive Cancer Gene Therapy. Genes 2019; 10(8), 560. doi:10.3390/genes10080560.
7. Iacobas DA, Tuli N, Iacobas S, Rasamny JK, Moscatello A, Geliebter J, Tiwari RM. Gene master regulators of papillary and anaplastic thyroid cancer phenotypes. Oncotarget 2018; 9(2), 2410-2424. doi: 10. 18632/oncotarget.23417.
8. Iacobas DA, Iacobas S. Towards a Personalized Cancer Gene Therapy: A Case of Clear Cell Renal Cell Carcinoma. Cancer & Oncol Res 2017; 5(3): 45-52. DOI:10.13189/cor.2017.050301.
9. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133906