Abstract
Expansion of Circulating Tumor Cells (CTC), the metastatic seeds of cancer in the blood stream, holds great potential in clinical application, especially towards precision medicine. Given the relatively rare nature of CTCs, their culture remains to be a significant challenge. When developing technologies for CTC culture, there are key elements that need careful consideration, including the speed of culture, compatibility with downstream analysis, and the implementation of the technology into established clinical daily routines. Herein, we briefly discuss the implications of our recent report of an ultrathin filter for the capture and culture of circulating colon cancer cells.
Commentary
Today, cancer is the second leading cause of death, with about 9.6 million deaths globally in 2018 [1]. At the end of the 19th century, Paul Ehrlich contributed a milestone to cancer research by introducing chemotherapy as a promising tumor treatment approach [2]. Since then, cancer treatment has undergone tremendous advances, with chemotherapy still being a widely used cancer treatment method today, however, often associated with severe side effects [3]. Immunotherapy emerged in the 1980’s, and by activating or boosting the body’s own antitumor mechanisms, [4] it has proven to be one of the most promising cancer treatments, showing fewer off-target effects than chemotherapy [5]. Thanks to advances of the nucleic acid technology, tremendous amount of data can now be collected for each individual patient. Once such data can be properly stratified and related to functional studies, the nucleic acid based technologies will most likely pave the way for early diagnosis and hence allowing cancer treatment in early disease progression [6,7].
In many cases, the cancer heterogeneity counteracts a general treatment approach that benefits every patient to the same extent [8,9]. Precision medicine for cancer is therefore seen as a landmark in cancer therapy. For effective drug testing and down-stream cancer analysis, an ex vivo 3D tumor model comprised of cancer cells from the individual patient would be a valuable tool. One possible resource to obtain ex vivo tumor models from patients might be circulating tumor cells (CTCs). Although CTCs are considered to be the main source of metastasis, their clinical application remain uncertain. Essentially, the CTC number in blood of especially colorectal cancer patients can be extremely low with 10-9.ml-1 among hematologic blood cells [10]. This rareness of CTCs in blood hampers the efficiency by which they can be analyzed, and used for functional studies and treatment screening. Several methods to isolate CTCs from patient blood have been developed over the last two decades [11], with CellSearch® as the only FDA approved method to isolate and enumerate CTCs. In recent years, focus has turned from enumeration to expansion of CTCs. The establishment of stable CTC cell lines derived from patient blood has been demonstrated and important insights into cancer biology could be obtained [12-20]. A few leading studies either successfully applied negative depletion methods [12-14,19,20] or used microfluidic devices [15-18,21] to culture CTCs from patient blood. Although highly depending on cancer type and disease status, negative depletion methods suffer from low culture success rates (e. g. 5.9% [12], 8.6% [13], 8.7% [20] and 2% [14]). Since time is precious in cancer treatment, rapid expansion of captured CTCs is a critical factor for CTC culture technology. The aforementioned negative depletion studies showed culture time duration varying from 3-4 weeks [13,20] to several months [12,14,19] in order to obtain stable CTC cultures. On the other hand, microfluidic devices have shown higher culture success rates (e. g. 16.7% [18] and 19.6–59.3% [16]), and sufficient CTC expansion e. g. for drug testing within 2 weeks [16]. However, microfluidic devices often suffer from their inherent complexity which hampers their integration into clinical routines. Thus there is still a need for CTC culture technologies which enable fast expansion of CTCs and makes it possible to analyze the expanded tumor material in a way that can easily adapt to clinical practices.
Electrospinning has been recognized to be a versatile technology to produce unique nanostructures that mimic native extracellular matrix. Together with their high specific surface area, electrospun fibers could not only facilitate the capture but also the on-site culture of CTCs. Several studies have applied electrospinning for capturing CTCs. Ueki et al. developed an anti-EpCAM conjugated solution electrospun polystyrene (PS) mesh. Blood spiked with MCF-7 breast cancer cells was filtered through the PS fibers mesh via vacuum aspiration. Although cell capture was enhanced up to 30% with thickened mesh of 100 um, no further culture of captured cells could be demonstrated [22]. Recently, Liu et al. demonstrated solution electrospun poly-(lactic-co-glycolic acid) (PLGA) fibers conjugated with anti-EpCAM and anti-N-Cadherin, for the capture of both epithelial and mesenchymal CTCs [23]. Here, MCF- 7 and GIST882, a gastrointestinal stromal tumor line representing a more mesenchymal-like cancer cell line, upon spike-in blood samples were incubated on top of the fibers for 40 min before immunochemistry. A capture efficiency of 60% could be achieved, however, without further culture of captured cells [23]. Xu et al. developed a microfluidic chip incorporated with hyaluronic acid functionalized electrospun PLGA fibers, able to isolate CD44 positive cancer cells. Additionally, it was shown that in principle captured cells could also be cultured directly on the device. However, an extremely high cancer cell concentration of 100,000 cells/ml, not clinical relevant, was used for perfusion culture experiments and no patient derived blood was used in this study. Culture under static conditions was demonstrated with 7000 HeLa cells per well. Possibly, culture experiments with lower cancer cell concentrations were also conducted, however, no clinical relevant cancer cell concentration for culture is clearly stated [24].
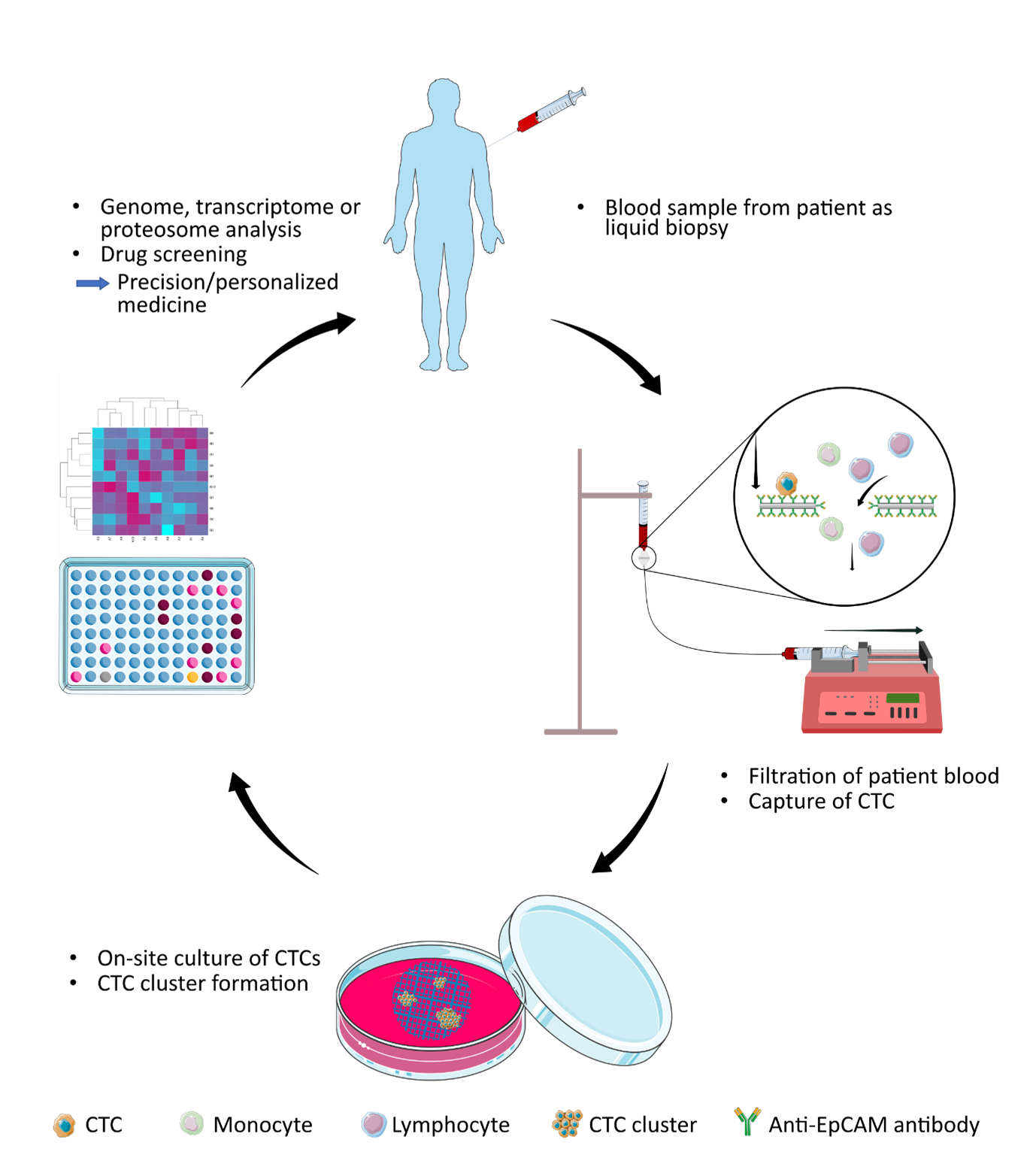
Advancing electrospinning with computer aided design additive technology, melt electrospinning written (MEW) of polymer melts with lower conductivity is distinguished by its robustness to produce tailored porosity, mechanical property, and transparency which facilitate both filtration and on-site cell culture/monitoring/down-stream analysis. Recently, our group firstly demonstrated a MEW fabricated polycaprolactone (PCL) filter conjugated with anti-EpCAM antibodies for the purpose of capturing and in situ expansion of epithelial CTCs [25]. We have shown, that as few as 20 HT29 colon cancer cells spiked into 4 ml (5 cells per ml) of whole blood can be captured and expanded in situ to tumor bodies visible to the naked eye within two weeks that can be microscopically monitored on-site followed with down-stream genomic/proteomic analysis. In perspective, this approach could facilitate precision medicine for cancer patients, by providing a platform for capture of CTCs from patient blood and further on-site culture to create cancer clusters within 2-4 weeks. Figure 1 illustrates the potential workflow and how the filter could be implemented in clinic, leading to a more individualized cancer treatment. We demonstrate, that our filter is highly compatible with down-stream analysis on both protein and nucleic acid level. The filter is only 10 μm thick, has a porosity of 60%, and immunochemistry-based analysis of the filtered cells is straightforward.
Figure 1. The CTC capture and culture approach put in perspective. A blood sample is drawn from a cancer patient as liquid biopsy. The blood sample is then filtrated using the anti-EpCAM conjugated melt electrospun filter. The close up view of how an EpCAM positive CTC is binding to the anti-EpCAM conjugated filter surface while white blood cells like monocytes and lymphocytes are passing the pores. After 2-4 weeks of on-site culture the tumor clusters could be used for further down-stream analysis like genome sequencing and drug screening.
Challenges and Future Outlook
Through tremendous efforts, technologies to isolate CTCs from patient blood samples for ex vivo culture have been developed. For all the technologies there is a tradeoff between capture efficiency and purity, which appears to be almost inversely related. Furthermore, it is a major challenge to make devices that allow for efficient capture and at the same time present an environment for the tumor cells where they will proliferate. Finally, the technology needs to be compatible with microscopy as this allows monitoring the capture and culture on-site. Microscopy is also a key step in the pathological analysis of the cancer disease. The characterization by immunochemistry is still and probably will continue to be a very important ground for the clinical treatment decision making. Indeed, the access to the tumor material after culture is one of the great advantages of the filter compared to some of the recently reported elegant microchip systems for CTC expansion [26,27].
One of the main arguments for choosing a filtration setup to expand CTCs, is the apparent simplicity of implementing the technology into the clinic. Filtration setups are intuitively easy to understand, and filtrations can be done in any standard laboratory facilities and do not require highly trained laboratory staff. Furthermore, our MEW fabricated filter profits from its low cost in production. MEW is a stable and cost-effective method to print PCL microfibers. The equipment necessary to conduct the filtration can be found in most laboratories (tubes, syringes and syringe pump). The simplicity of the filtration setup together with the on-site culture/ monitoring/downstream analysis features stand out in comparison to multicomponent microfluidic devices [21,28].
A filter such as the one reported herein can enable both affinity and size-based capture of CTCs. Indeed, by being able to tune pore sizes, flow dynamics, and the number of different conjugated antibodies, a broader range of CTCs can be captured [11,29,30]. Furthermore, although suggested to facilitate cancer cluster formation [16], too high amounts of WBCs might decrease the CTC culture efficiency. Hence, aiming for a pore size distribution allowing more WBC to pass the filter pores while maintaining a high CTC capture efficiency could increase chances to successfully expand captured CTCs. WBCs are known to be less stiff due to lower N/C ratio compared to CTCs [31] and might be able to be pushed through the smaller pores by a higher flowrate.
Clearly the culture conditions for captured CTCs will have a major impact on the ability of CTCs to proliferate and form clusters. It is known that not all CTCs have the potential to proliferate to form new cancerous colonies, since some CTCs might lack proliferative features and maintain in a state of dormancy without cancer outgrowth, or are simply dead [32]. Dormant CTCs might require external signals for further proliferation like growth factors and cytokines [32]. Among those, hypoxia has been reported to play a role as an external signal for cancer outgrowth, where cancer cells become more invasive and tend to more metastatic behavior [33]. It has been reported that hypoxia can act as an external cue stimulating dormant cells to exhibit a more stem cell-like phenotype and ultimately leading to cancer outgrowth [34]. Erler et al. showed the importance of hypoxia in establishing a premetastatic niche where CTCs might reside and form a metastatic colony [35]. Since oxygen levels in outgrowing tumors can reach below 2%, future culture experiments under hypoxia condition could mimic the hypoxic environment the captured CTC might originate from [36].
However, the interplay between CTC dormancy and hypoxia remains elusive [37], and CTCs have been successfully cultured under both hypoxic [13,15] and nonhypoxic condition [12]. A few other studies suggest that hypoxia might act suppressive on CTCs in dormant states. It has been shown that the hypoxia-inducible gene domain family member 1A (HIGD1A), a hypoxia induced survival factor, can also promote a state of dormancy in cancer cells [38]. Additionally, a study by Hofstetter et al. suggested that hypoxia is increasing protein phosphatase 2A activity, a factor important for cell cycle regulation and growth inhibition [39].
Culture under low attachment and low serum levels [15] has been suggested to be essential for successful culture, whereas others suggest co-culture [17] to be an important parameter. This underscores that there might not be onefits- all standard for culturing these very heterogenous cancer cells.
Our technology of capturing and culturing CTCs using melt electrowritten filter will be validated by patientderived blood samples. If successful, we have developed a platform for CTC isolation and rapid ex vivo cultivation, which allows many crucial downstream analysis methods, and can be easily implemented in established clinical routines.
References
2. DeVita Jr VT, Rosenberg SA. Two hundred years of cancer research. New England Journal of Medicine. 2012 Jun 7;366(23):2207-14.
3. Schirrmacher V. From chemotherapy to biological therapy: A review of novel concepts to reduce the side effects of systemic cancer treatment. International Journal of Oncology. 2019 Feb 1;54(2):407-19.
4. Dobosz P, Dzieciatkowski T. The intriguing history of cancer immunotherapy. Frontiers in Immunology. 2019 Dec 17;10:2965.
5. Riley RS, June CH, Langer R, Mitchell MJ. Delivery technologies for cancer immunotherapy. Nature Reviews Drug Discovery. 2019 Mar;18(3):175-96.
6. Xiang D, Shigdar S, Qiao G, Wang T, Kouzani AZ, Zhou SF, et al. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: the next generation of cancer medicine. Theranostics. 2015;5(1):23.
7. Hassan EM, DeRosa MC. Recent advances in cancer early detection and diagnosis: Role of nucleic acid based aptasensors. TrAC Trends in Analytical Chemistry. 2020 Mar 1;124:115806.
8. Strickaert A, Saiselet M, Dom G, De Deken X, Dumont JE, Feron O, et al. Cancer heterogeneity is not compatible with one unique cancer cell metabolic map. Oncogene. 2017 May;36(19):2637-42.
9. Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nature Reviews Clinical Oncology. 2018 Feb;15(2):81.
10. Cayrefourcq L, Mazard T, Joosse S, Solassol J, Ramos J, Assenat E, et al . Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Research. 2015 Mar 1;75(5):892-901.
11. Joosse SA, Gorges TM, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Molecular Medicine. 2015 Jan;7(1):1-1.
12. Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014 Sep 25;159(1):176-87.
13. Grillet F, Bayet E, Villeronce O, Zappia L, Lagerqvist EL, Lunke S, et al. Circulating tumour cells from patients with colorectal cancer have cancer stem cell hallmarks in ex vivo culture. Gut. 2017 Oct 1;66(10):1802-10.
14. Zhao P, Zhou W, Liu C, Zhang H, Cheng Z, Wu W, et al. Establishment and Characterization of a CTC Cell Line from Peripheral Blood of Breast Cancer Patient. Journal of Cancer. 2019;10(24):6095-6104.
15. Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014 Jul 11;345(6193):216-20.
16. Khoo BL, Grenci G, Jing T, Lim YB, Lee SC, Thiery JP, et al. Liquid biopsy and therapeutic response: Circulating tumor cell cultures for evaluation of anticancer treatment. Science Advances. 2016 Jul 1;2(7):e1600274.
17. Zhang Z, Shiratsuchi H, Lin J, Chen G, Reddy RM, Azizi E, et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget. 2014 Dec;5(23):12383.
18. Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014 Jul 11;345(6193):216-20.
19. Soler A, Cayrefourcq L, Mazard T, Babayan A, Lamy PJ, Assou S, et al. Autologous cell lines from circulating colon cancer cells captured from sequential liquid biopsies as model to study therapy-driven tumor changes. Scientific Reports. 2018 Oct 29;8(1):1-12.
20. Brungs D, Minaei E, Piper AK, Perry J, Splitt A, Carolan M, et al. Establishment of novel long-term cultures from EpCAM positive and negative circulating tumour cells from patients with metastatic gastroesophageal cancer. Scientific Reports. 2020 Jan 17;10(1):1-13.
21. Goudar VS, Yeh PH, Wu SY, Chu CH, Lu LS, Yang CH, et al. Live circulating tumour cells selection on digitized self-assembled cell array (Digi-saca) chip by in-parallel/insitu image analysis, cell capture, and cultivation. Sensors and Actuators B: Chemical. 2020 Apr 11:128002.
22. Ueki T, Yoshihara A, Teramura Y, Takai M. Fast and selective cell isolation from blood sample by microfiber fabric system with vacuum aspiration. Science and Technology of Advanced Materials. 2016 Dec 1;17(1):807-15.
23. Liu H, Wang Z, Chen C, Ding P, Sun N, Pei R. Dualantibody Modified PLGA Nanofibers for Specific Capture of Epithelial and Mesenchymal CTCs. Colloids and Surfaces B: Biointerfaces. 2019 Sep 1;181:143-8.
24. Xu G, Tan Y, Xu T, Yin D, Wang M, Shen M, et al. Hyaluronic acid-functionalized electrospun PLGA nanofibers embedded in a microfluidic chip for cancer cell capture and culture. Biomaterials Science. 2017;5(4):752- 61.
25. Jørgensen ML, Müller C, Sikkersoq M, Nadzieja M, Zhang Z, Su Y, e t al. A melt-electrowritten filter for capture and culture of circulating colon cancer cells. Materials Today Bio. 2020 May 1:100052.
26. Khoo BL, Lee SC, Kumar P, Tan TZ, Warkiani ME, Ow SG, et al. Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget. 2015 Jun 20;6(17):15578.
27. Zhang Z, Shiratsuchi H, Lin J, Chen G, Reddy RM, Azizi E, et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model.Oncotarget. 2014 Dec;5(23):12383-97.
28. Khoo BL, Grenci G, Lim YB, Lee SC, Han J, Lim CT. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nature Protocols. 2018 Jan;13(1):34-58.
29. Satelli A, Brownlee Z, Mitra A, Meng QH, Li S. Circulating Tumor Cell Enumeration with a Combination of Epithelial Cell Adhesion Molecule–and Cell-Surface Vimentin–Based Methods for Monitoring Breast Cancer Therapeutic Response. Clinical Chemistry. 2015 Jan 1;61(1):259-66.
30. Kowalik A, Kowalewska M, Gózdz S. Current approaches for avoiding the limitations of circulating tumor cells detection methods—implications for diagnosis and treatment of patients with solid tumors. Translational Research. 2017 Jul 1;185:58-84.e15.
31. Bagnall JS, Byun S, Begum S, Miyamoto DT, Hecht VC, Maheswaran S, et al. Deformability of tumor cells versus blood cells. Scientific Reports. 2015 Dec 18;5:18542.
32. Gao XL, Zhang M, Tang YL, Liang XH. Cancer cell dormancy: mechanisms and implications of cancer recurrence and metastasis. OncoTargets and Therapy. 2017;10:5219.
33. Al Tameemi W, Dale TP, Al-Jumaily RM, Forsyth NR. Hypoxia-modified cancer cell metabolism. Frontiers in Cell and Developmental Biology. 2019 Jan 29;7:4.
34. Weidenfeld K, Schif-Zuck S, Abu-Tayeh H, Kang K, Kessler O, Weissmann M, et al. Dormant tumor cells expressing LOXL2 acquire a stem-like phenotype mediating their transition to proliferative growth. Oncotarget. 2016 Nov 1;7(44):71362.
35. Erler JT, Bennewith KL, Cox TR, Lang G, Bird D, Koong A, et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009 Jan 6;15(1):35-44.
36. Jing X, Yang F, Shao C, Wei K, Xie M, Shen H, et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Molecular Cancer. 2019 Dec 1;18(1):157.
37. Butturini E, Carcereri de Prati A, Boriero D, Mariotto S. Tumor dormancy and interplay with hypoxic tumor microenvironment. International Journal of Molecular Sciences. 2019 Jan;20(17):4305.
38. Ameri K, Jahangiri A, Rajah AM, Tormos KV, Nagarajan R, Pekmezci M, et al. HIGD1A regulates oxygen consumption, ROS production, and AMPK activity during glucose deprivation to modulate cell survival and tumor growth. Cell Reports. 2015 Feb 17;10(6):891-9.
39. Hofstetter CP, Burkhardt JK, Shin BJ, Gürsel DB, Mubita L, Gorrepati R, et al. Protein phosphatase 2A mediates dormancy of glioblastoma multiforme-derived tumor stem-like cells during hypoxia. PloS one. 2012 Jan 11;7(1):e30059.