Abstract
Among clinical and industrial important microorganisms Streptomyces are the largest source of natural products. But only a fraction of all Biosynthetic Gene Clusters in their genome is active under laboratory conditions. During the last years research has clarified the role of cyclic dimeric 3`-5` guanosine monophosphate, c-di-GMP, as an ubiquitous secondary messenger. It is not only a regulator of morphological development but also of the production of natural products. This makes it an interesting starting point for the development of new natural products. The production and degradation of c-di-GMP builds up the first layer of regulation. A set of enzymes with redundant or at least overlapping function is responsible for these reactions. Subsequently several effectors such as BldD or σWhiG affect the expression of many target genes. Through countless interactions within this system - comprising feedback loops and bilateral regulation - a multilayered system makes it possible to have a finely regulated system that can react quickly to external and internal environments. In this system c-di-GMP helps to provide BldD its function as the so called “master regulator” in Streptomyces. Interfering in this system with genetic manipulations such as knockouts or overexpression of genes led to altering levels of c-di-GMP. Thus, it was possible to change the production levels of produced natural products and activate silent gene clusters.
Keywords
c-di-GMP, Streptomyces, Natural products, Antibiotics, Second messengers
Abbreviations
c-di-GMP: Cyclic dimeric 3`-5` Guanosine Monophosphate; CSR: Cluster Situated Regulator; BGC: Biosynthetic Gene Cluster; DGC: Diguanylate Cyclase; GAF: Mammalian cGMP-regulated PDEs, Anabaena adenylyl cyclases and Escherichia coli transcription activator FhlA; PAC: Photoactivated Adenylyl Cyclase; PAS: Per-Arnt-Sim; PDE: Phosphodiesterase.
Introduction
Natural products play a crucial role in the development of drugs. Over the last forty years one third of all approved drugs are natural products or derivatives from them. Additionally, another third uses at least a pharmacophore of a natural product. The therapeutic areas do not only cover the widely known fields of antibiotics and cytostatics, but also anticoagulants, anti-hypertensive or anti-diabetic drugs and many more [1]. Nevertheless, antibiotics are one of the most prominent fields for natural products. Increasing antibiotic resistance in the upcoming years is leading to an urgent need for the development of new antibiotics [2]. The World Health Organization stated the antibiotic resistance crisis to be a “global public health concern”. Also, the Center for Disease Control and Prevention and the European Medicines Agency are substantially concerned about the course in the last decades [2-4]. Streptomyces, a genus of the family Streptomycetaceae and the class Actinobacteria, are the largest source of natural products among microorganisms. A large proportion of all antibiotics originally derive from them [ 5]. However, in the last years only a handful of new drugs in the antibiotic field were approved. So, there is an urgent need to search for new natural compounds beyond of the existing ones [6]. A high rate of rediscovered compounds is one problem of common techniques for screening for new natural products with antibiotic effect [7]. Consequently, new techniques to discover completely new entities should be searched and used. Streptomyces have this potential because a large percentage of Biosynthetic Gene Clusters (BGC) remains inactive under laboratory conditions. Thus, undiscovered natural products are burrowed within their genome. Analysis using antiSMASH revealed that each sequenced Streptomyces genome has 36.5 BGCs on average [8], while producing only single natural product. Additionally, it is estimated that only 1 % of all Actinobacteria are cultivable and can be identified by using standard methods [9]. To use Streptomyces to their full potential we need techniques to activate more biosynthetic gene clusters. C-di-GMP is a secondary messenger involved in crucial pathways for natural products biosynthesis. Biotechnological interventions in this overarching regulation circuit are able to enhance yields and furthermore activate biosynthetic gene clusters as described below.
Streptomyces
Streptomyces are gram-positive, non-motile, soil-dwelling Actinobacteria, with a high G+C content genome. They are multicellular bacteria and undergo a complex life cycle [5,10,11]. The germination starts with a suitable trigger. From the unigenomic spore a germ tube emerges, which grows mainly at the tip. Through branching a tightly interwoven mycelium is formed and quasi-exponential growth is possible.Additionaly speta are formed in older parts of the mycelium [12-14]. After a few days the colonies begin to form aerial hyphae, which themselves mainly grow at their tips. The life cycle is summarized in Figure 1. A set of genes is involed in this process. According to the appearance of the respective knockout mutants the genes were called “bld” after their “bald” look on agar plates. The long, often curled aerial hyphae undergo multiple synchronus cell devisions to form spores. Responsible for this is another set of genes called “whi” after their knockout mutants, which are unable to produce pigmented spores and therefore look white [15-17]. These names bld and whi may be irritating because of the intricate effects of some enzymes. BldD does not only have an influence on the formation aerial hyphae but also plays a role in spore formation through interactions with WhiB and ssgR [18]. The names were given years ago just for phenotypic looks, but the underlying mechanisms were not elucidated yet.
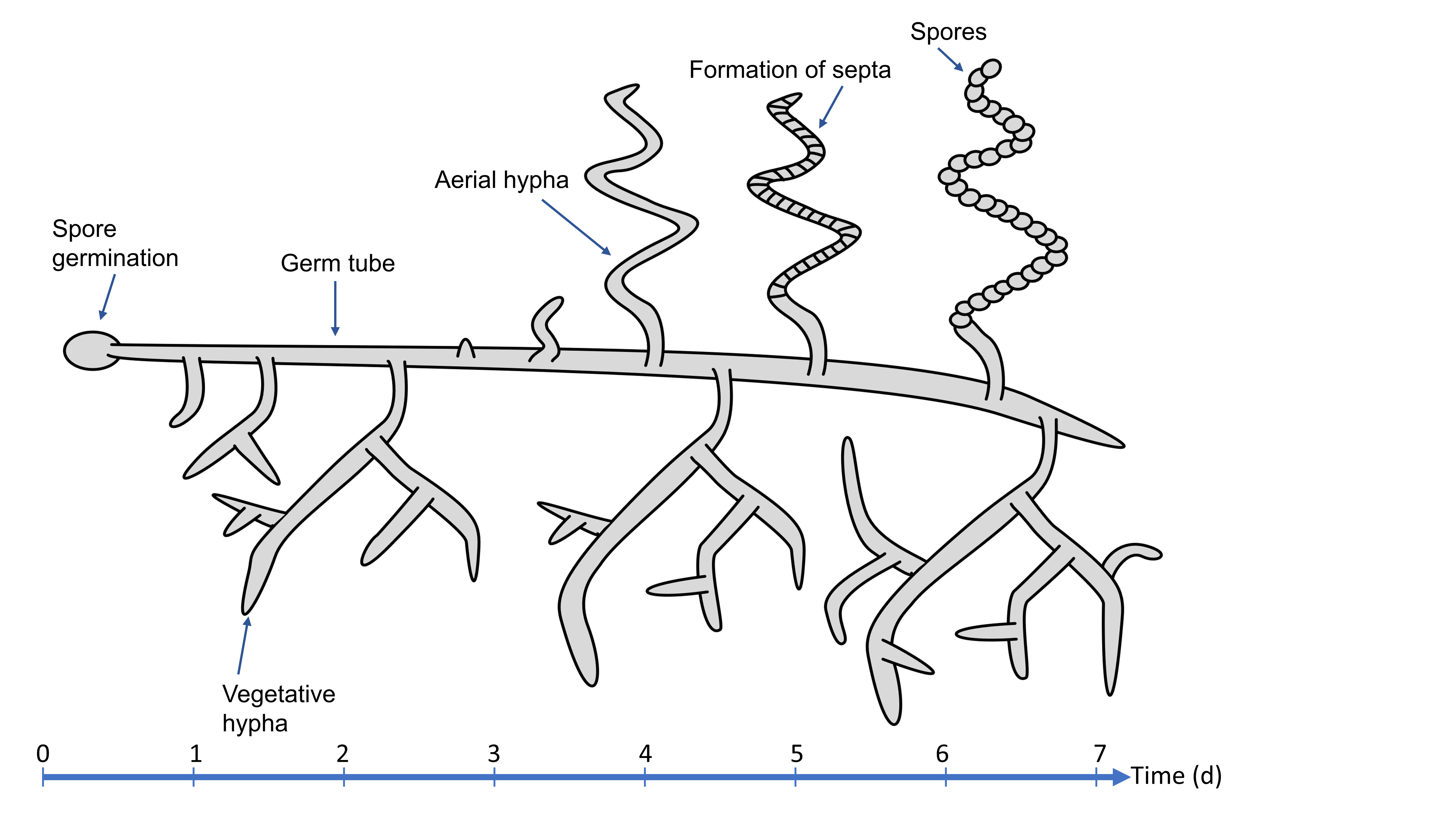
Figure 1: Lifecycle of Streptomyces. Development from a single spore into vegetative hyphae, aerial hyphae and formation of spores.
The genus Streptomyces was first proposed by Waksman und Henrici [19]. In the following years they began to isolate compounds such as streptothricin and actinomycin from S. antibioticus and S. lavendulae. Since the discovery of penicillin approximately 70,000 natural products from microbials have been identified of which 20,000 are from Actinobacteria [9,20]. The natural products have a large structural diversity as shown in Figure 2. The figure shows some of the most important natural products derived from Streptomyces.
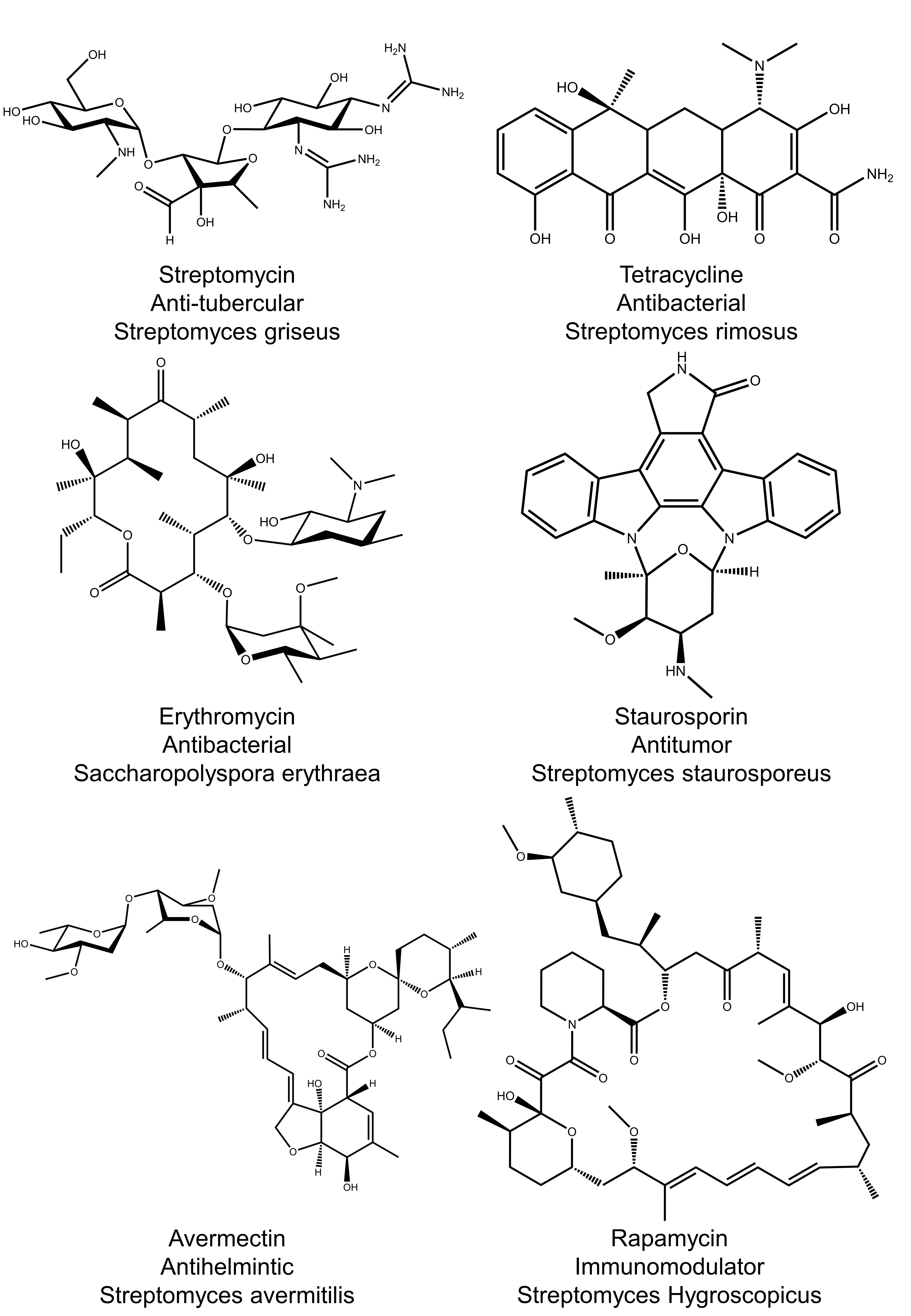
Figure 2: Structural diversity of natural products produced by the named strains. The description shows the name of the compound, the medical use, and the originating strain. Streptomycin is one of the first compounds isolated from Streptomyces. Ivermectin is a semisynthetic derivative from Avermectin. The others show different structure types occurring as natural products of Streptomyces.
The synthesis of natural products in Streptomyces is organized in biosynthetic gene clusters. These clusters are 20 kb to over 100 kb in size and contain most of the essential genes for biosynthesis [21]. Recent genome sequencing efforts coupled with advances in bioinformatics indicate that the majority of biosynthetic gene clusters are not expressed under normal laboratory conditions. Termed 'silent' or 'cryptic', these gene clusters represent a treasure trove for discovery of novel small molecules, their regulatory circuits and their biosynthetic pathways [22,23]. A lot of different techniques have been deployed to generate compounds from these silent gene clusters. For example, co-cultivation with other microorganisms to mimic interactions between species or applying chemical elicitors with metabolome analysis revealed new compounds [24-27]. One problem with these approaches is that they are governed to an analytical scale.
That is why we take a look at a global technique to activate new biosynthetic gene clusters or increase amounts of produced natural products through intervention in a higherlevel, pleiotropic regulator. For this it is crucially important to not only be able to detect the gene clusters in the genomic sequence, but also to know regulatory cascades of their activation. In most of the BGCs so-called cluster sited regulators (CSRs) are located directly within the biosynthetic genes. They are necessary for transcription or activity of the other genes. But some gene clusters do not have any CSRs and the number of so-called “CSR-free” BGCs is growing through advances in sequencing.
It is important to note that a regulator within a BGC can have an effect on more than one BGC, meaning that it has a more global function. Additionally, there are global regulators governing production of natural products and also morphological development. The current state of research will be described in the following chapters.
Originally the strain S. coelicolor was used as a model organism and was also the first fully sequenced genome of all Streptomyces [28]. It was and is the best overall studied strain. Until now almost 20 different secondary metabolites of this strain were analyzed. Among others are the polyketide actinorhodin and the red tripyrrole prodiginines [29,30]. Recently, S. venezuelae, producer of chloramphenicol and the polyketide jadomycin, is emerging as a new model strain. Because it also sporulates in liquid culture, it is considerably more suitable for the evaluation of developmental processes [31,32]. This is especially true, because surface grown colonies, which are necessary for S. coelicolor to undergo the full development circle, are compromised by developmental asynchrony and heterogeneity making statements about regulons difficult. Because some Streptomyces strains are extremely difficult to handle, it is easier to express the biosynthetic gene cluster of interest in a heterologous host. Well studied strains like S. coelicolor, S. lividans, S. albus and S. avermiltilis have been used and also specifically modified for this purpose by deleting their BGCs to minimize interactions [ 33,34]. Other strains from the so called “rare Actinobacteria” (for example Saccharopolyspora erythraea) are used because of the interesting natural products they are producing.
In particular the investigations of the direct effects of c-di- GMP were performed with the two model organisms S. coelicolor and S. venezuelae [35-38]. Recently also a different, S. ghanaensis, known for the production of moenomycin, an antibiotic which was used for decades as a feedstuff without rising resistance levels [39], was also investigated [11,40,41].
The second messenger C-di-GMP
Cyclic dimeric 3´-5´ guanosine monophosphate (c-di- GMP) was first identified in 1987 as an allosteric activator of a bacterial cellulose synthase in the alphaproteobacterium Gluconacetobacter xylinus. It took quite a while until the first signs did show the impact of c-di-GMP. Ten more years were needed for the characterization of the main domains of the first response regulators in Caulobacter crescentus and Bordetella pertussis [42,43]. These domains are conserved throughout different species. Later it was shown, that the amino acid sequence GGDEF, in the so-called GGDEF-domain, has diguanylate cyclase (DGC) activity, building up c-di-GMP out of two molecules of GTP [43]. In contrast to this the EAL amino acid sequence, in the EAL-domain and also the HD-GYP domain, are catalyzing the degradation of c-di-GMP in pGpG and subsequently into GMP and therefore has the function of a phosphodiesterase (PDE) [5,44–46]. The c-di-GMP turnover is summarized in Figure 3.
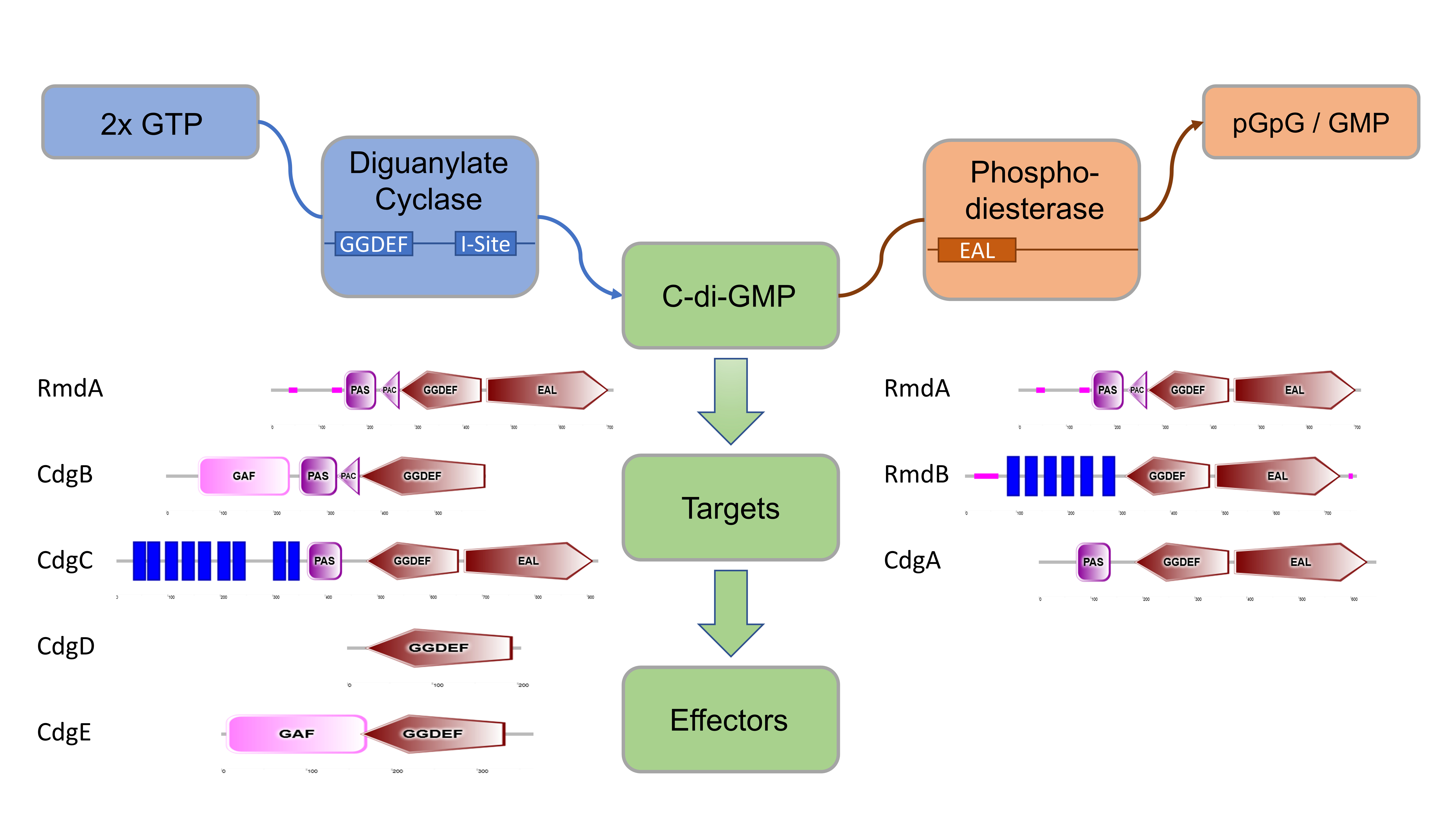
Figure 3: Overview of c-di-GMP turnover. C-di-GMP is produced by DGCs out of two molecules of GTP and degraded by PDEs into pGpG or GMP. In its active form it forms complexes with target proteins, which in turn regulate the expression of the effectors. The schemes of the enzymes listed are from S. ghanaensis [47].
It is important to note that some of the enzymes carry both necessary domains within their sequence, but in most cases only one of them is active. The other one is degenerate in the surrounding of the active site and does not carry out its original function. In some rare cases it was shown that both domains are still active and the protein can carry out both reactions. Their activity is mediated by stimuli that alter the activity of either one of the domains [48].
In the following years its role was enlarged to a ubiquitous secondary messenger in bacteria.
C-di-GMP is responsible for different kinds of changes in morphology like transition from sessility to motility in Salmonella enterica, Pseudomonas aeruginosa and Escherichia coli [49,50]. Associated with sessility is also the biofilm formation [51]. Another function of c-di-GMP is to modulate pathogeny in among others Yersinia pestis and Vibrio cholerae [52–54].
The downstream effectors are very diverse reaching from PilZ domains [55,56] to riboswitches [57]. In Streptomyces two special complexes carry out the main function, which are described below.
About ten years ago first research about the involvement of c-di-GMP in Streptomyces was published [10]. At first the research in Streptomyces focused on the impact of c-di-GMP on morphological development. During the past years, additionally the effects on the production of secondary metabolites were analyzed more detailed [11,36,40,41,58]. Each Streptomyces strain has a certain set of multiple enzymes, varying from five to twelve enzymes in total in all sequenced Streptomyces [5,38,59]. Some of these enzymes like CdgA, CdgB and RmdB are existing in almost every strain, while others are present in only a few [5].
C-di-GMP is able to integrate a wide range of different internal and external signals through numerous additional domains. Three of these sensory and regulatory domains play the main role in Streptomyces: PAS (Per-Arnt-Sim)/ PAC (Photoactivated adenylyl cyclase) and GAF (mammalian cGMP-regulated PDEs, Anabaena adenylyl cyclases and Escherichia coli transcription activator FhlA) [40]. PAS is a common signal sensing domain, with various ligands, such as heme, FMN or FAD, mainly present in archea and prokaryotes. They can occur both intracellularly and extracellularly. In S. ghanaensis an ortholog of CdgC was shown to possess a heme, potentially sensing signals from O2, Co or NO [40]. Maybe analogous to a PAS domain from E. coli, which showed oxygen dependent activity in the past [60]. GAF domains are also small molecule binding domains. They are also found in mammalian phosphodiesterases with cGMP as one of the known ligands [61,62]. In Streptomyces and in a lot of bacteria these domains have not been analyzed in detail, meaning that a lot of uncertainty exists about potential ligands and their influence on the activity of the enzymes [5]. For better understanding how changes in intracellular c-di- GMP levels impact the production of secondary metabolites we need to take a look at the effectors of c-di-GMP first.
C-di-GMP effectors and targets
C-di-GMP is the active secondary messenger. In its main functions it is building complexes with enzymes, which subsequently bind their target DNA sequences. These targets play roles in different pathways and build a multilayered system of regulation to be able to adapt to different internal and external effects [63]. BldD was proven to create a complex out of four molecules of c-di-GMP and two molecules of BldD bind to their signature DNA-sequence (5`-TNAC[N]5GTNA-3´) via their DNA binding domain. The conserved amino acid sequence motif RXD-X8-RXXD binds c-di-GMP [35,36]. When enough c-di-GMP is present the complex is formed and binds the DNA. First it was shown that the binding represses the transcription of the regulon. Just recently in 2019 also a mode of c-di-GMP dependent activation of genes was published [64]. Studies on the regulation and the interaction revealed that in S. coelicolor 261 Genes were changed over twofold in expression through the knockout of bldD. Some of the genes are involved in life cycle development or secondary metabolites production, while other proteins still have unknown functions [10]. The resulting effects on secondary metabolites vary from complete stop to enhancement of production between different strains and the underlying causes remain to be elucidated [10,41,64].
One important target of BldD is BldA. BldA is the only tRNA in the genome of Streptomyces that can translate the UUA-Codon [65,66]. Because of the high GC-content of approximately 72%, this codon is used in only 2% of all genes, which still amounts to 145 genes. None of these genes is crucial for survival, so a bldA null mutant is still viable. Also, most of these genes are not conserved throughout Streptomyces. Some of them are in species specific BGCs for example. This makes it hard to predict general effects for BldA. The amount of BldA can be a limiting factor for the translation of a protein and proceeding the production of a natural product. Almost 50% of Streptomyces with partially dysfunctional bldA gene produced a new compound after restoring its function. This is not only due to direct presence of a UUA codon in a BGC but also indirect effects through regulators containing the codon [67,68].
AdpA, also known as BldH, is an important regulator of core promoters within BGCs and furthermore one of the most versatile transcription factors in Streptomyces and contains a conserved TTA codon [23,69,70]. This global regulation is particularly important in CSR-free BGCs [23]. The activity of AdpA is also regulated by RNase III, AbsB (Antibiotic biosynthesis gene B), by cleavage of its mRNA and through direct interaction with BldD. The knockout of absB in S. ghanaensis more than doubled the production of moenomycin whereas the knockout of adpA completely abolished the production [70].
The second important direct target of c-di-GMP is the complex between σWhiG and RsiG, which is only formed in the presence of c-di-GMP. It was first analyzed in S.venezuelae but is conserved throughout all Streptomyces. The σWhiG/RsiG complex binds two molecules of c-di-GMP with two conserved E(X)3S(X)2R(X)3Q(X)3D domains. At lower intracellular levels of c-di-GMP σWhiG is released of the complex and can subsequently bind an RNA polymerase. The consensus sequence for DNA binding of the RNA polymerase and σWhiG is very similar, but not identical to the phylogenetically related sigma factors for flagellum biosynthesis [71,72]. The transcription of whiG itself is regulated inhibitory directly through BldD and enhancing through WblA. For the latter it is unclear whether the interaction is direct or indirect [10,73]. The two main targets of σWhiG are WhiH and WhiI. These two on the other hand, affect more than 100 genes involved in late sporulation [72]. The WhiH protein has similarities to GntR family transcription factors, a family with other members known to be involved in developmental regulation, including DevA and DasR [74-76]. Until now only a few connections between σWhiG and the production of secondary metabolites have been made. In S. chattanoogensis L10 σWhiG binds to CSR within the natamycin BGC. Additionally, indirect effects on pathway specific positive regulators were shown [77]. How this indirect regulation is enabled remains unclear. In contrast to that in the same strain a second BGC was downregulated and in S. coelicolor the overexpression of σWhiG leads to inhibition of the production of actinorhodine [77,78]. In the future, it will be interesting to find out through which regulatory elements σWhiG controls the production of secondary metabolites.
One possibility would be, that it is achieved through the WhiB-like (Wbl) regulators, a family of transcription factors. They were shown to have an influence in both sporulation and secondary metabolites production. The knockout of the most abundant member, wblA, led to a more than 10-fold increase in the production of tiancimycins in S. sp. CB03234 [79]. The function of WblA is a transcription repressor, nevertheless also activation and dual repressor activation have been reported [80]. In S. ansochromogenes 7100 and S. somaliensis SCSIO ZH66 the inactivation of wblA led to activation of cryptic BGCs. In both studies, a BGC was inactivated in addition to the one being activated for unknown reason [73,81]. The knockout also almost abolished the expression of several other Whi and Wbl genes. Which of these effects are direct and which are indirect still needs to be investigated [73]. WblA is negatively regulated through BldD [41] but contrary results have been observed for the interaction between AdpA and WblA. In the model strain S. coelicolor and in S. ghanaensis AdpA negatively regulates wblA transcription [41,82] on the other hand in S. griseus and S. chattanoogensis L10, AdpA shows a positive regulation on WblA [83,84]. In both cases direct interactions with the promotor region have been shown. Also, the overexpression of wblA led to increased production of natamycin in S. chattanoogensis L10, contrary to the results mentioned before [84].
Furthermore, there is a complex of BldM, regulated by BldD, and WhiI, which is regulated by WhiG [10,18]. The fact alone that BldM used to be called WhiK shows that it is under heavy regulation and is interwoven in multiple pathways [85]. bldM is under direct control of BldD and additionally the sigma factor directing bldM transcription, BldN, is also under control of BldD [10,18]. BldM has two different DNA binding motifs, one is a homo-dimer, the other one is a hetero-dimer with BldM and WhiI. WhiI is one of the main targets of WhiG and is connected to the BldD pathway again. The targets of the complexes simultaneously occur during the development of aerial hyphae and in sporulation [18]. Although the influence of BldM on morphological development is clear, it remains unclear whether there is an additional link to antibiotic production. Because the knockout of BldM is leading to a “bald” morphology there is a connection to genes responsible for the formation of aerial hyphae [18,85]. Further research is needed to evaluate whether this connects to natural product regulation pathways. The pathways described above are shown in Figure 4.
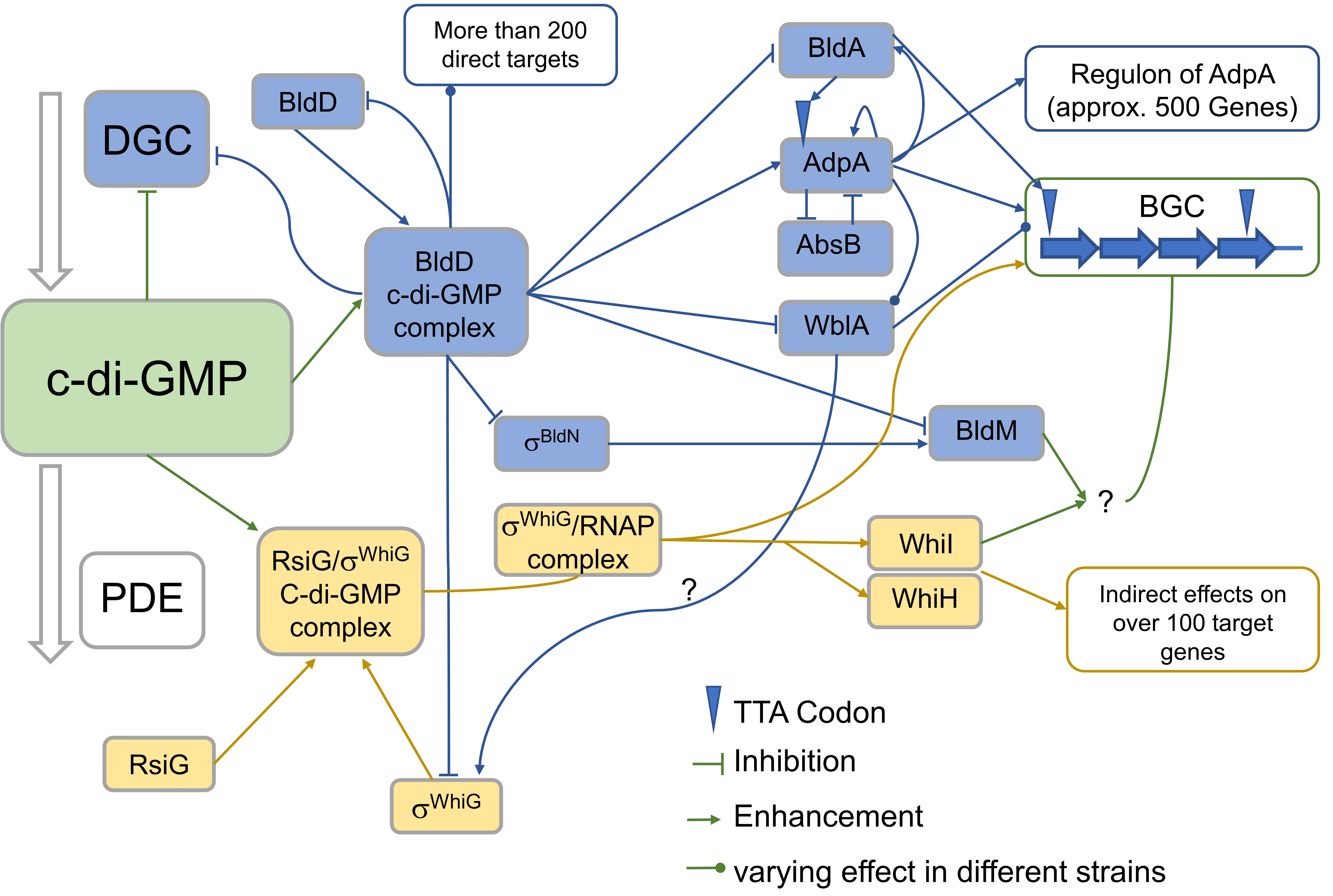
Figure 4: Overview of the most important known interactions of c-di-GMP and their downstream effectors. The effect is only shown if it is a conserved interaction. For example, BldA has more individual interactions in certain strains if they contain other TTA-codons. The effects of the BldM-WhiI complex are not yet analyzed in regards to natural products synthesis. AdpA and WblA show different effects on each other in different strains. WblA also showed activating and inhibiting effects on BGCs. It has to be analyzed whether the effect of WblA and σWhiG is conserved in Streptomyces.
Influence of changes in synthesis and degradation on effectors of c-di-GMP
In recent years it was shown that not only changes on the effector levels are a possibility to influence the production of natural products. In several strains, as discussed above, knockout and overexpression studies were conducted on DGCs and PDEs [38]. However, most of them focused on the morphologic influence of c-di-GMP and not the production of secondary metabolites [5,15,86]. Only recently, first works on this topic have been published [11,38,41].
The idea was that by changing levels of c-di-GMP, it could be possible to alter the activity of the respective complexes with BldD and σWhiG /RsiG and subsequently the expression of genes relevant for natural products biosynthesis. A knockout of a DGC and accordingly the overexpression of a PDE is supposed to lower the levels of c-di-GMP and consecutively leads to the breakdown of the complexes. In the case of BldD the disintegration of the complex is preventing the DNA binding. In the case of σWhiG /RsiG the breakdown releases σWhiG , which is then able to recruit an RNA polymerase on its respective target genes. The exact opposite is assumed for a knockout of a PDE and overexpression of a DGC [11,37]. This is a simplified view, not taking into account that there are potential feedback loops or enzymes with overlapping function impairing the impact on the level of c-di-GMP [15,40,87]. Liu et al. listed an overview on the effects in S. venezuelae and S. coelicolor with contrary effects on the production of actinorhodin in different S. coelicolor DGC mutants [38]. The formation of the complexes of c-di-GMP with BldD and σWhiG /RsiG is supposed to prevent the transition into aerial hyphae and sporulation respectively. Until now it remains still unclear whether the new findings of BldD enhancing the transcription are playing an important role in this regard [64]. But it would support the fact, that the overexpression of different DGC enzymes leads to different results in terms of natural products production. Another unclarity arises from studies from S. ghanaensis. In this strain opposing to the findings in S. coelicolor, the enzymatic activity of CdgA was a PDE and not a DGC, although they share high amino acid identity [11]. This means it is not possible to predict the activity of these enzymes just from their overall sequence similarity. On the other hand, it is possible to see degenerate domains, when crucial residues are missing [11]. Further evaluations in S. ghanaensis revealed that the knockout of rmdB led to the activation of cryptic BGCs. Two new natural products are produced. The same procedure in S. albus led also to the production of new compounds [41].
Table 1 summarizes knockouts, overexpression experiments and enzyme activities of S. coelicolor, S. venezuelae and S.ghanaensis and their impact on natural products synthesis [ 5,10,11,15,38,40,41,59,86].
| Strain | Production of SMs | |||
|---|---|---|---|---|
| Protein | Target of BldD | Enzym Activity | Overexpression | Knockout |
| S. ghanaensis | ||||
| CdgA | PDE | ↓ | ↑ | |
| CdgB | x | DGC* | ↑ | ↓ |
| CdgC | x | DGC | ↑ | ↓ |
| CdgD | - | ↓ | ↑ | |
| CdgE | DGC | → | → | |
| RmdA | Bifunctional | → | ↑ | |
| RmdB | x | PDE | ↓ | ↑ |
| S. venezuelae | ||||
| CdgA | x | DGC* | - | - |
| CdgB | x | DGC* | - | ↓ |
| CdgC | x | DGC | - | ↓ |
| CdgD | DGC* | - | - | |
| CdgE | x | DGC* | - | - |
| RmdA | PDE [residual DGC activity] | - | → | |
| RmdB | PDE | - | → | |
| S. coelicolor | ||||
| CdgA | x | DGC* | ↓ | - |
| CdgB | x | DGC | ↓ | → |
| CdgC | / | DGC* | ↑ | - |
| CdgD | / | DGC | ↓ | - |
| CdgE | DGC* | - | - | |
| RmdA | PDE | - | - | |
| RmdB | PDE | - | - | |
Table 1: Overview of c-di-GMP turnover enzymes in S. ghanaensis, S. coelicolor and S. venezuelae. Target of BldD means, that a BldD binding site is located in the promoter region of the respective gene. Arrows show the effect of overexpression and knockout on the production of secondary metabolites. “-“ means that it was not yet analyzed. * = no in vitro validation. CdgB does not contain an EAL domain, so it is not able to have PDE activity.
Conclusion and Outlook
Cyclic-di-GMP has demonstrated to be an interesting starting point for the generation of new natural products. It was shown that through changes in the effector proteins as well as through changes in the turnover proteins it was possible to activate cryptic gene clusters.
C-di-GMP metabolizing enzymes are the upstream targets in this complex and interwoven pathway. The underlying cause why Streptomyces have multiple enzymes with redundant function is not yet clarified. A possible explanation would be, that they are expressed in a time-dependent manner and have different tasks throughout the development of Streptomyces enzymes [5,40]. Alternatively, the global pool of c-di-GMP is not the central point, but local pools and modules of enzymes play their role in the regulatory pathways [5,15]. This is in contradiction to the fact, that overexpression of other enzymes diminishes the effect of a knockout of redundant ones. But it is safe to say that redundant enzymes have different localizations, through for example transmembrane domains. This also makes it possible that external stimuli are integrated into the cells through extracellular sensing domains [88]. Additionally, intercellular proteins with their own domains are able to integrate other signals. But little is known about the natural ligands and their effect on the activity of most of these domains.
Further analyses are required, if the activation of cryptic BGCs - as shown above - is a general principle or if it was a hit by chance. In case, it can be confirmed, it will be a profound tool for the generation of new natural products of Streptomyces in general. This is especially true, because it is a conserved pathway and specifically rmdB occurs in more than 90 % of all Streptomyces.
Furthermore, it will be interesting to see which modulators are involved to connect c-di-GMP turnover enzymes and the activation of BGCs. Hundreds of enzymes are in the regulon of c-di-GMP effectors, but which are the decisive ones? This implies also an advantage the turnover enzymes as a target point: Not only single effectors are influenced, but the whole regulon. There is to note that this also can have adverse impact, because it is not precisely targeted.
Declarations of Interest
None.
References
2. Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645–58.
3. Michael CA, Dominey-Howes D, Labbate M. The antimicrobial resistance crisis: Causes, consequences, and management. Front Public Heal. 2014 Sep 16;2(SEP):145.
4. Spellberg B, Srinivasan A, Chambers HF. New societal approaches to empowering antibiotic stewardship. Vol. 315, JAMA - Journal of the American Medical Association. American Medical Association; 2016. p. 1229–30.
5. Al-Bassam MM, Haist J, Neumann SA, Lindenberg S, Tschowri N. Expression Patterns, Genomic Conservation and Input Into Developmental Regulation of the GGDEF/EAL/HD-GYP Domain Proteins in Streptomyces. Front Microbiol. 2018;9:2524.
6. Silver LL. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24(1):71–109.
7. Safaei N, Mast Y, Steinert M, Huber K, Bunk B, Wink J. Angucycline-like Aromatic Polyketide from a Novel Streptomyces Species Reveals Freshwater Snail Physa acuta as Underexplored Reservoir for Antibiotic-Producing Actinomycetes. Antibiot (Basel, Switzerland). 2020 Jan 1;10(1):1–13.
8. Lee N, Hwang S, Kim W, Lee Y, Kim JH, Cho S, et al. Systems and synthetic biology to elucidate secondary metabolite biosynthetic gene clusters encoded in: Streptomyces genomes. Vol. 38, Natural Product Reports. 2021. p. 1330–61.
9. Bérdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. Vol. 65, Journal of Antibiotics. Nature Publishing Group; 2012. p. 385–95.
10. Den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol. 2010;(78):361–79.
11. Nuzzo D, Makitrynskyy R, Tsypik O, Bechthold A. Identification and Characterization of Four c-di-GMP-Metabolizing Enzymes from Streptomyces ghanaensis ATCC14672 Involved in the Regulation of Morphogenesis and Moenomycin A Biosynthesis. Microorganisms. 2021;9(2).
12. Hopwood DA, Tobias K, Bibb MJ, Buttner MJ, Chater KF. Practical Streptomyces Genetics. John Innes Foundation; 2000. 613 p.
13. Chater KF. Genetics of differentiation in Streptomyces. Vol. 47, Annual Review of Microbiology. 1993. p. 685–713.
14. Kelemen GH, Buttner MJ. Initiation of aerial mycelium formation in Streptomyces. Curr Opin Microbiol. 1998;1(6):656–62.
15. Hull TD, Ryu M-H, Sullivan MJ, Johnson RC, Klena NT, Geiger RM, et al. Cyclic Di-GMP phosphodiesterases RmdA and RmdB are involved in regulating colony morphology and development in Streptomyces coelicolor. J Bacteriol. 2012;194(17):4642–51.
16. Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, et al. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 2003 Jul 15;17(14):1727–40.
17. Bush MJ, Bibb MJ, Chandra G, Findlay KC, Buttner MJ. Genes required for aerial growth, cell division, and chromosome segregation are targets of whia before sporulation in Streptomyces venezuelae. MBio. 2013 Sep 24;4(5).
18. Al-Bassam MM, Bibb MJ, Bush MJ, Chandra MJ, Buttner GJ. Response Regulator Heterodimer Formation Controls a Key Stage in Streptomyces Development. PLoS Genet. 2014;10(8):1004554.
19. Waksman SA, Henrici AT. The Nomenclature and Classification of the Actinomycetes. J Bacteriol. 1943 Oct;46(4):337–41.
20. Katz L, Baltz RH. Natural product discovery: past, present, and future. J Ind Microbiol {\&} Biotechnol. 2016;43(2–3):155–76.
21. Nah HJ, Pyeon HR, Kang SH, Choi SS, Kim ES. Cloning and heterologous expression of a large-sized natural product biosynthetic gene cluster in Streptomyces species. Front Microbiol. 2017 Mar 15;8(MAR):394.
22. Okada BK, Seyedsayamdost MR. Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol Rev. 2017 Jan 1;41(1):19–33.
23. Makitrynskyy R, Ostash B, Tsypik O, Rebets Y, Doud E, Meredith T, et al. Pleiotropic regulatory genes bldA, adpA and absB are implicated in production of phosphoglycolipid antibiotic moenomycin. Open Biol. 2013;3(10):130121.
24. Xu F, Nazari B, Moon K, Bushin LB, Seyedsayamdost MR. Discovery of a Cryptic Antifungal Compound from Streptomyces albus J1074 Using High-Throughput Elicitor Screens. J Am Chem Soc. 2017 Jul 12;139(27):9203.
25. Bertrand S, Bohni N, Schnee S, Schumpp O, Gindro K, Wolfender JL. Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnol Adv. 2014;32(6):1180–204.
26. Marmann A, Aly AH, Lin W, Wang B, Proksch P. Co-cultivation- -a powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar Drugs. 2014;12(2):1043–65.
27. Rosen PC, Seyedsayamdost MR. Though Much Is Taken, Much Abides: Finding New Antibiotics Using Old Ones. Biochemistry. 2017 Sep 19;56(37):4925–6.
28. Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 2002 May 9;417(6885):141–7.
29. Rudd BAM, Hopwood DA. Genetics of actinorhodin biosynthesis by Streptomyces coelicolor A3(2). J Gen Microbiol. 1979;114(1):35–43.
30. Chater KF, Dyson PJ. Recent advances in understanding Streptomyces. F1000Research. 2016;5(2795).
31. Bibb MJ, Domonkos Á, Chandra G, Buttner MJ. Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by σ(BldN) and a cognate anti-sigma factor, RsbN. Mol Microbiol. 2012 Jun;84(6):1033–49
32. Chater KF. Recent advances in understanding Streptomyces [version 1; referees: 4 approved]. Vol. 5, F1000Research. F1000 Research Limited; 2016. p. 2795.
33. McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides. Science (80- ). 1993;262(5139):1546–50.
34. Ahmed Y, Rebets Y, Estévez MR, Zapp J, Myronovskyi M, Luzhetskyy A. Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters. Microb Cell Fact. 2020 Jan 9;19(1):1–16.
35. Schumacher MA, Zeng W, Findlay KC, Buttner MJ, Brennan RG, Tschowri N. The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex. Nucleic Acids Res. 2017;45(11):6923–33.
36. Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, et al. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell. 2014;158(5):1136–47.
37. Haist J, Neumann SA, Al-Bassam MM, Lindenberg S, Elliot MA, Tschowri N. Specialized and shared functions of diguanylate cyclases and phosphodiesterases in Streptomyces development. Mol Microbiol. 2020 Nov 1;114(5):808–22.
38. Liu X, Zheng G, Wang G, Jiang W, Li L, Lu Y. Overexpression of the diguanylate cyclase CdgD blocks developmental transitions and antibiotic biosynthesis in Streptomyces coelicolor. Sci China Life Sci. 2019 Nov 1;62(11):1492–505.
39. Lopatniuk M, Ostash B, Makitrynskyy R, Walker S, Luzhetskyy A, Fedorenko V. Testing the utility of site-specific recombinases for manipulations of genome of moenomycin producer Streptomyces ghanaensis ATCC14672. Journal of Applied Genetics. 2015 Nov;56(4):547-50.
40. Nuzzo D, Makitrynskyy R, Tsypik O, Bechthold A. Cyclic di-GMP cyclase SSFG_02181 from Streptomyces ghanaensis ATCC14672 regulates antibiotic biosynthesis and morphological differentiation in streptomycetes. Sci Rep. 2020;10:12021.
41. Makitrynskyy R, Tsypik O, Nuzzo D, Paululat T, Zechel DL, Bechthold A. Secondary nucleotide messenger c-di-GMP exerts a global control on natural product biosynthesis in streptomycetes. Nucleic Acids Res. 2020;48(3):1583–98.
42. Merkel TJ, Barros C, Stibitz S. Characterization of the bvgR locus of Bordetella pertussis. J Bacteriol. 1998;180(7):1682–90.
43. Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, et al. Three cdg Operons Control Cellular Turnover of Cyclic Di-GMP in Acetobacter xylinum: Genetic Organization and Occurrence of Conserved Domains in Isoenzymes. J Bacteriol. 1998;180(17):4416.
44. Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and Characterization of a Cyclic di-GMP-specific Phosphodiesterase and Its Allosteric Control by GTP. J Biol Chem. 2005;280(35):30829–37.
45. Tchigvintsev A, Xu X, Singer A, Chang C, Brown G, Proudfoot M, et al. Structural insight into the mechanism of cyclic di- GMP hydrolysis by EAL domain phosphodiesterases. J Mol Biol. 2010;402(3):524.
46. Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005 Jul;187(14):4774–81.
47. Letunic I, Khedkar S, Bork P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Research. 2021 Jan 8;49(D1):D458-60.
48. Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Vol. 7, Nature Reviews Microbiology. 2009. p. 724–35.
49. Simm R, Morr M, Kader A, Nimtz M, Römling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004 Aug 1;53(4):1123–34.
50. Hecht GB, Newton A. Identification of a novel response regulator required for the swarmer-to- stalked-cell transition in Caulobacter crescentus. J Bacteriol. 1995;177(21):6223–9.
51. Karaolis DKR, Rashid MH, Chythanya R, Luo W, Hyodo M, Hayakawa Y. c-di-GMP (3'-5'-Cyclic Diguanylic Acid) Inhibits Staphylococcus aureus Cell-Cell Interactions and Biofilm Formation. Antimicrob Agents Chemother. 2005 Mar;49(3):1029.
52. Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol. 2004 Oct;54(1):75–88.
53. Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004 Aug;53(3):857.
54. Tischler AD, Camilli A. Cyclic Diguanylate Regulates Vibrio cholerae Virulence Gene Expression. Infect Immun. 2005 Sep;73(9):5873.
55. Christen M, Christen B, Allan MG, Folcher M, Jenö P, Grzesiek S, et al. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci U S A. 2007 Mar 6;104(10):4112.
56. Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153.
57. Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, et al. Riboswitches in Eubacteria Sense the Second Messenger Cyclic Di-GMP. Science. 2008 Jul 18;321(5887):411.
58. Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–73.
59. Tschowri N. Cyclic Dinucleotide-Controlled Regulatory Pathways in Streptomyces Species. J Bacteriol. 2016;198(1):47–54.
60. Kitanishi K, Kobayashi K, Kawamura Y, Ishigami I, Ogura T, Nakajima K, et al. Important roles of Tyr43 at the putative heme distal side in the oxygen recognition and stability of the Fe(II)-O2 complex of YddV, a globin-coupled heme-based oxygen sensor diguanylate cyclase. Biochemistry. 2010 Dec 14;49(49):10381–93.
61. Hurley JH. GAF domains: cyclic nucleotides come full circle. Vol. 2003, Science’s STKE : signal transduction knowledge environment. 2003.
62. Schmidt HHHW, Hofmann F, Stasch J-P. Handbook of Experimental Pharmacology, Volume 191: cGMP: Generators, Effectors and Therapeutic Implications. Vol. 190, Handbook of Experimental Pharmacology. 2009. p. 93–107.
63. Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol. 2017;15(5):271–84.
64. Yan H, Lu X, Sun D, Zhuang S, Chen Q, Chen Z, et al. BldD, a master developmental repressor, activates antibiotic production in two Streptomyces species. Mol Microbiol. 2020 Jan 1;113(1):123– 42.
65. Kwak J, McCue LA, Kendrick KE. Identification of bldA mutants of Streptomyces griseus. Gene. 1996;171(1):75–8.
66. Lawlor EJ, Baylis HA, Chater KF. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1987;1(10):1305–10.
67. Hackl S, Bechthold A. The Gene bldA, a Regulator of Morphological Differentiation and Antibiotic Production in Streptomyces. Arch Pharm (Weinheim). 2015 Jul 1;348(7):455–62.
68. Gessner A, Heitzler T, Zhang S, Klaus C, Murillo R, Zhao H, et al. Changing Biosynthetic Profiles by Expressing bldA in Streptomyces Strains. ChemBioChem. 2015 Oct 1;16(15):2244–52.
69. Higo A, Horinouchi S, Ohnishi Y. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol Microbiol. 2011 Sep;81(6):1607–22.
70. Xu W, Huang J, Lin R, Shi J, Cohen SN. Regulation of morphological differentiation in S. coelicolor by RNase III (AbsB) cleavage of mRNA encoding the AdpA transcription factor. Mol Microbiol. 2010;75(3):781–91.
71. Schumacher MA, Gallagher KA, Holmes NA, Chandra G, Henderson M, Kysela DT, et al. Evolution of a σ-(c-di-GMP)-anti-σ switch.
72. Gallagher KA, Schumacher MA, Bush MJ, Bibb MJ, Chandra G, Holmes NA, et al. c-di-GMP Arms an Anti-σ to Control Progression of Multicellular Differentiation in Streptomyces. Mol Cell. 2020 Feb 6;77(3):586-599.e6.
73. Huang H, Hou L, Li H, Qiu Y, Ju J, Li W. Activation of a plasmidsituated type III PKS gene cluster by deletion of a wbl gene in deepsea-derived Streptomyces somaliensis SCSIO ZH66. Microb Cell Fact. 2016 Jun 27;15(1).
74. Persson J, Chater KF, Flärdh K. Molecular and cytological analysis of the expression of Streptomyces sporulation regulatory gene whiH. FEMS Microbiol Lett. 2013 Apr 1;341(2):96–105.
75. Hoskisson PA, Rigali S, Fowler K, Findlay KC, Buttner MJ. DevA, a GntR-like transcriptional regulator required for development in Streptomyces coelicolor. J Bacteriol. 2006 Jul;188(14):5014–23.
76. Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, et al. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008 Jul;9(7):670–5.
77. Liu SP, Yu P, Yuan PH, Zhou ZX, Bu QT, Mao XM, et al. Sigma factor WhiGch positively regulates natamycin production in Streptomyces chattanoogensis L10. Appl Microbiol Biotechnol. 2015 Feb 28;99(6):2715–26.
78. Mendez C, Chater KF. Cloning of whiG, a gene critical for sporulation of Streptomyces coelicolor A3(2). J Bacteriol. 1987;169(12):5715–20.
79. Zhang F, Gao D, Lin J, Zhu M, Zhuang Z, Duan Y, et al. Construction of Inducible Genetic Switch for the Global Regulator WblA to Sustain Both Overproduction of Tiancimycins and On- Demand Sporulation in Streptomyces sp. CB03234. ACS Synth Biol. 2020 Jun 19;9(6):1460–7.
80. Huang X, Ma T, Tian J, Shen L, Zuo H, Hu C, et al. wblA, a pleiotropic regulatory gene modulating morphogenesis and daptomycin production in Streptomyces roseosporus. J Appl Microbiol. 2017;123(3):669–77.
81. Lu C, Liao G, Zhang J, Tan H. Identification of novel tylosin analogues generated by a wblA disruption mutant of Streptomyces ansochromogenes. Microb Cell Fact. 2015 Nov 2;14(1).
82. Lee HN, Kim JS, Kim P, Lee HS, Kim ES. Repression of antibiotic downregulator WblA by AdpA in Streptomyces coelicolor. Appl Environ Microbiol. 2013 Jul;79(13):4159–63.
83. Higo A, Hara H, Horinouchi S, Ohnishi Y. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 2012 Jun;19(3):259–73.
84. Yu P, Liu S-P, Bu Q-T, Zhou Z-X, Zhu Z-H, Huang F-L, et al. WblA ch , a Pivotal Activator of Natamycin Biosynthesis and Morphological Differentiation in Streptomyces chattanoogensis L10, Is Positively Regulated by AdpA ch. Appl Environ Microbiol. 2014;80(22):6879–87.
85. Molle V, Buttner MJ. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol Microbiol. 2000;(6):1265–78.
86. Tran NT, Hengst CDD, Gomez-Escribano JP, Buttner MJ. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J Bacteriol. 2011 Jun;193(12):3100–8.
87. Chou SH, Galperin MY. Diversity of cyclic di-GMP-binding proteins and mechanisms. Vol. 198, Journal of Bacteriology. 2016. p. 32–46.
88. Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the First 25 Years of a Universal Bacterial Second Messenger. Microbiol Mol Biol Rev. 2013 Mar;77(1):1.
