Commentary
Protein ubiquitination is a major post-translational mechanism that regulates fate and function of many proteins in the cell, either by regulating their abundance by the 26S-proteasome-ubiquitin system or by modulating protein activity by the attachment of the ubiquitin modifier [1]. Thus, in addition to targeting proteins for proteasomal degradation, ubiquitin plays a role in many other non-degradative processes in the cell, including transcription, cell cycle, DNA repair, apoptosis, immune response, endosomal sorting, among others [2]. Protein ubiquitination requires the coordination of an enzymatic cascade composed by the E1 activation enzyme, E2 conjugating enzyme, and the E3 ligase. Sometimes, E2 can directly guide ubiquitin to bind to the substrate, but in most circumstances, the cooperative work of E3 ligase is required for catalysis and specificity [3]. Deubiquitinating enzymes (DUBs) can reverse ubiquitination by removing ubiquitin from protein targets having an opposite function to the E3 ligases. Coordination of these two activities contributes to the fine-tune regulation of target ubiquitination inside the cell, and in some cases their abundance or half-life when ubiquitin targets protein degradation by the proteasome. Deregulation of DUBs activity can be linked to several diseases, such as cancer or neurodegeneration among others, indicating the major role of this proteolytic activity for a correct cellular homeostasis.
The “ubiquitinome” includes all types of ubiquitin chains that can be formed by conjugation on the different lysines in the ubiquitin surface, including the N-terminal residue of ubiquitin and the combination with other ubiquitin-like proteins, such as SUMO (Small-Ubiquitin Modifier). K48- linked chains target proteins for proteasomal degradation, and K63-linked chains is a non-degradative signal that plays a role in DNA damage repair; both are perhaps the most common type of poly-Ub chains. Whereas there are over 800 types of ubiquitin E3 ligases, which contribute to the formation of the large variety of ubiquitin chains, the DUBs family (DeUBiquitinating enzymes) contains over 90 members, also contributing to the regulation of the “ubiquitinome”.
The DUBs family is composed by 6 structural classes of proteolytic enzymes according to their structure, including the ubiquitin-specific proteases (USP), the ovarian tumor proteases (OTU), the ubiquitin C-terminal hydrolases (UCHs), the motif interacting with ubiquitin containing novel DUB family (MINDYs), the JAB1/MPN/ MOV34 metalloprotease DUBs (JAMMs), and ubiquitinlike proteases (ULPs) [1]. In addition to the conserved catalytic domain, DUBs usually contain adjacent domains often involved in target specificity. This is particularly relevant in the USP family, the most abundant class of DUBs that displays a large variety of additional domains N- or C-terminally to the catalytic domain, in some cases involved in substrate-binding, contributing to ubiquitin chain-type selectivity. USP25 is an important member of the USP family and a positive regulator in the Wnt/β- catenin signaling pathway [4]. The discovery of USP25’s auto-tetramer catalytic suppression mechanism attracted much more attention in the field.
We and others have recently revealed a novel and unique mechanism of regulation of the USP25 activity, which depends on the 180 amino acid insert in the middle of the USP catalytic domain [5-7]. USP25 can display two quaternary structures in vitro and in vivo: dimer or activated enzyme; and tetramer or inhibited enzyme. In the tetramer conformation an inhibitory loop from the central insertion domain partly occludes the active site substrate surface, thus inhibiting the USP25 activity (Figure 1). Switching between these two quaternary assemblies possibly regulates the activity of USP25 inside the cell. This tetramer auto-inhibitory mechanism of regulation in USP25 has no parallel in any other member of the DUB family [5]. Intriguingly, despite its high sequence homology with USP28 (more than 50% of sequence identity), structural and functional analyses reveal a different mechanism of activity regulation for USP28 [6,7]. Both dimer structures of USP25 and USP28 are quite similar and both rely on the coiled-coil helical central domain (Figure 1), however they have significant differences in the catalytic activity in vitro, and the efficiency of USP28 is 6 times that of USP25 [6].
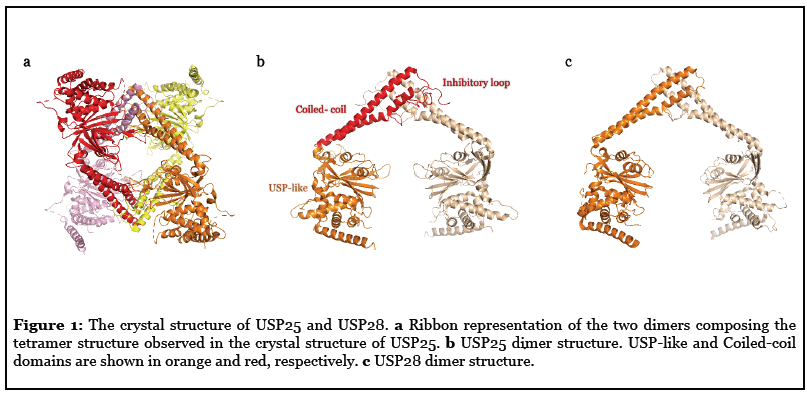
So far in the USP family, no deubiquitinating enzymes other than USP25 have been found to have this tetrameric auto-inhibitory function, but there are other examples with auto-inhibition situations, such as in USP14. According to cryo-electron microscopy and related structural studies, it has been found that the autoinhibited USP14 apoenzyme has a low catalytic activity, but after binding to the proteasome, USP14 is activated and its activity considerably increased [8-10]. Besides, the active site of USP7 can switch between active and inactive state through interaction with its C-terminal ubiquitin-like domain. The active structure involves a rearrangement of the catalytic domain of USP7 and increases the activity by 100 times [11,12]. A similar allosteric regulation also occurs in USP4. The DUSP-Ubl domain at the N-terminal of USP4 is an essential part of the full catalytic activity of USP4. DUSP promotes the release of ubiquitin by interfering with the mechanism, thereby accelerating catalytic conversion [13]. Similarly, Ubp8 singly is a catalytically inactive and needs to be allosterically activated by other subunits of the SAGA complex [14,15].
Most USPs are multidomain enzymes containing a conserved catalytic domain of nearly 300 amino acids, structurally portrayed as palm, thumb, and fingers, that contain all elements needed for the proteolytic activity [16]. The adjacent domains of USPs are usually involved in their particular functions, sometimes regulating its activity by binding protein substrates and other modulators. The central catalytic domains of USP25 and USP28 share 57% sequence identity and both contain an insertion domain of approximately 180 amino acids, formed by a long coiled-coil structure, which acts as a dimerization hub, followed by an unstructured loop, which is responsible of the tetramer in USP25 (Figure 1) [5-7]. The dimer structures in USP25 and USP28 show a similar “double head” structure, exposing two catalytic active sites but with slightly different orientations, which probably prevents the formation of the tetramer assembly in USP28. In USP25, the loop connecting the coiled-coil insertion domain interacts with the USP domain from the opposite dimer, allowing the formation of a tetramer (or “dimer of dimers”) [5]. However, the equivalent loop in USP28, which displays a lower sequence homology in the USP28 family, does not allow the formation of a tetramer assembly [6]. Interestingly, a chimeric construct of USP28 containing the loop insertion of USP25 cannot form a tetramer [7], indicating that the correct orientation of the USP catalytic domains in the dimer structure is essential for the correct assembly of the tetramer. We named the loop connecting the coiled-coil insertion domain as “inhibitory loop” because by forming the tetramer assembly it occludes the binding surface of the “proximal” ubiquitin to the USP25 active site. So, despite the high degree of sequence homology between USP25 and USP28, they display distinct regulatory mechanisms: both exist as active dimers, but only USP25 can also exist as an inhibited-tetramer.
To the question why both USP25 and USP28 have evolved as active dimers over monomers, it could be speculated that USP25 or USP28 in the dimeric state would facilitate substrate binding by interacting to two ubiquitin sites of a substrate simultaneously. Such coordination of the deubiquitinating activity from the two active sites of USP25 or USP28 might increase its proteolytic efficiency by directly or indirectly binding to proteins complexes [6,7]. In fact, several USP28 substrates adopt a dimeric state, and the elongated shape of the tankyrase substrate of USP25 possesses at least two ubiquitination sites [17,18], which might be optimal for the “double head” activity of the USP25 and USP28 dimers. We can also speculate whether similar oligomerization domain inserts may exist in other proteases, allowing them to switch between monomer, dimer or higher oligomerization states.
In the case of USP25, a major question arises to whether such mechanism oscillating between these two oligomeric states occurs in the cell. We and others have shown the presence of dimer and tetramer USP25 assemblies in vitro and in vivo. It could be easily conceived an activation mechanism by which binding to the substrate triggers the tetramer to dimer transition, however, at least in vitro binding to ubiquitin chains does disrupt the tetramer [6]. We have proposed a mechanism by which a posttranslational modification, such as phosphorylation, could trigger the oligomerization switch between dimer and tetramer. In fact, a single mutation of Tyr454 for glutamic acid (a phosphomimetic mutation), located at the coiled-coil dimerization domain interface, impedes the formation of the tetramer in vitro, in contrast to the mutation for phenylalanine, which forms a stable tetramer alike wild-type USP25 [5]. Others speculate about other post-translational modifications, such a cis-trans prolylisomerase that could convert the tetrameric cis Pro535 to trans Pro, causing the dissociation of the tetramer [6]. It would be interesting in the future to investigate the molecular mechanism that shifts the equilibrium between active or inactive state of USP25.
Abnormal activation of Wnt signaling pathway plays a crucial role in the development of human cancer. The expression of USP25 regulates Wnt signaling by controlling the levels of tankyrases and Axin [4], thus USP25 may be considered to function as an oncogene. Besides, it has been recently discovered that USP25 can prevent the degradation of BCR-ABL protein and ensure the proliferation of Phpositive leukemia cells, which may become a new target for the development of chronic myelogenous leukemia (CML) drugs [19]. Another important function recently assigned to USP25 entails a role in host defense against RNA and DNA viruses. After being infected by RNA or DNA viruses, USP25 can actively regulate virus-triggered IRF3 and NF- κB activation and subsequent induction of type I IFN and pro-inflammatory cytokines by stabilizing TRAF3 and TRAF6, respectively, thus the DUB activity of USP25 can be considered as an innate immune response against DNA and RNA viruses [20]. Also, another report indicates that after viral infection or lipopolysaccharide (LPS) treatment, the expression level of USP25 is significantly up-regulated [21]. Recently, chloroquine (CQ) has been considered an effective treatment for COVID-19, but causing a widespread controversy. In the treatment of malaria, CQ disrupts the growth of parasites by increasing the pH of food vacuoles, but in the antiviral process, the mechanism of CQ has not been fully determined, but recent results also indicate that the antiviral effect of CQ may be caused by the alkalization of phagolysosomes or endolysosomes [22]. Interestingly, since CQ increases the expression of USP25 mRNA and protein in a dose-dependent manner [23], perhaps in the process of using CQ to treat COVID-19, USP25 might play a role in the regulation of endolysosomes pathway. It will be interesting to follow future research on this matter.
In summary, the discovery of the auto-inhibitory mechanism of USP25 has revealed new molecular insights into its regulatory mechanism, which can help to develop new tools to tackle connected pathologies and develop drugs of potential clinical significance.
Acknowledgments
Y.L. and B.L. acknowledge their scholarship to the China Scholarship Council program from the Chinese government. D.R. acknowledges the funding form the Spanish Government (PGC2018-098423-B-I00).
References
2. Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001 Jul 27;106(2):145-55.
3. Streich FC Jr, Lima CD. Structural and functional insights to ubiquitin-like protein conjugation. Annu Rev Biophys. 2014;43:357-79.
4. Xu D, Liu J, Fu T, Shan B, Qian L, Pan L, et al. USP25 regulates Wnt signaling by controlling the stability of tankyrases. Genes Dev. 2017 May 15;31(10):1024-1035.
5. Liu B, Sureda-Gómez M, Zhen Y, Amador V, Reverter D. A quaternary tetramer assembly inhibits the deubiquitinating activity of USP25. Nat Commun. 2018 Nov 26;9(1):4973.
6. Sauer F, Klemm T, Kollampally RB, Tessmer I, Nair RK, Popov N, et al. Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities. Mol Cell. 2019 May 2;74(3):421-435.e10.
7. Gersch M, Wagstaff JL, Toms AV, Graves B, Freund SMV, Komander D. Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity. Mol Cell. 2019 May 2;74(3):436-451.e7.
8. Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010 Sep 9;467(7312):179-84.
9. Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, et al. Structure and mechanisms of the proteasomeassociated deubiquitinating enzyme USP14. EMBO J. 2005 Nov 2;24(21):3747-56.
10. Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, et al. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci U S A. 2015 Jul 14;112(28):8626-31.
11. Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell. 2011 Oct 7;44(1):147-59.
12. Rougé L, Bainbridge TW, Kwok M, Tong R, Di Lello P, Wertz IE, et al. Molecular Understanding of USP7 Substrate Recognition and C-Terminal Activation. Structure. 2016 Aug 2;24(8):1335-1345.
13. Clerici M, Luna-Vargas MP, Faesen AC, Sixma TK. The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat Commun. 2014 Nov 18;5:5399.
14. Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010 May 21;328(5981):1025-9.
15. Köhler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell. 2010 May 14;141(4):606-17.
16. Ye Y, Scheel H, Hofmann K, Komander D. Dissection of USP catalytic domains reveals five common insertion points. Mol Biosyst. 2009 Dec;5(12):1797-808.
17. Eisemann T, McCauley M, Langelier MF, Gupta K, Roy S, Van Duyne GD, et al. Tankyrase-1 Ankyrin Repeats Form an Adaptable Binding Platform for Targets of ADPRibose Modification. Structure. 2016 Oct 4;24(10):1679- 1692.
18. Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011 Oct;10(10):M111.013284.
19. Shibata N, Ohoka N, Tsuji G, Demizu Y, Miyawaza K, Ui- Tei K, et al. Deubiquitylase USP25 prevents degradation of BCR-ABL protein and ensures proliferation of Ph-positive leukemia cells. Oncogene. 2020 May;39(19):3867-3878.
20. Lin D, Zhang M, Zhang MX, Ren Y, Jin J, Zhao Q, et al. Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6. Proc Natl Acad Sci US A. 2015 Sep 8;112(36):11324-9.
21. Ren Y, Zhao Y, Lin D, Xu X, Zhu Q, Yao J, et al. The type I interferon-IRF7 axis mediates transcriptional expression of Usp25 gene. Journal of Biological Chemistry. 2016 Jun 17;291(25):13206-15.
22. Carrière F, Longhi S, Record M. The endosomal lipid bis(monoacylglycerol) phosphate as a potential key player in the mechanism of action of chloroquine against SARS-COV-2 and other enveloped viruses hijacking the endocytic pathway. Biochimie. 2020 May 30: S0300- 9084(20)30129-2.
23. Ding C, Li F, Long Y, Zheng J. Chloroquine attenuates lipopolysaccharide-induced inflammatory responses through upregulation of USP25. Can J Physiol Pharmacol. 2017 May;95(5):481-491.
