Abstract
Long interspersed elements 1 (LINE-1 or L1) are currently the only active form of autonomous retrotransposons that exist in the human genome. The 6 kb L1 element encodes its own proteins to mediate retrotransposition without the aid of components from other retrotransposons. Moreover, L1 not only can transpose its’ own element but L1 proteins are also used for retrotransposition by other retrotransposons in the human genome including short interspersed elements (SINEs), of which Alu elements are a subset. L1 elements are expressed in the pluripotent cells of the early embryo and are then repressed in most somatic cells. They are, however, expressed in neurons in the brain and can become de-repressed in some cancer cells. However, the consequence of activity in these cells is not fully understood. In this review, we discuss the approach of using proteomic analysis to understand the function of the most abundant of the L1-derived proteins, ORF1p, in human embryonic stem cells (hESCs) and how we can apply proteomic approaches to decipher the role of L1-derived proteins in neurons and cancer cells. We will also discuss other recent findings about ORF1p, and proteins and RNAs with which it associates in mammalian cells and discuss how these findings affect our view of how this protein may impact human biology.
Introduction
In the last few decades there has been a growing interest in the role of transposable elements (TEs), colloquially referred to as “jumping genes” in human biology [1-4]. TEs, and a specific subset of this clade, retrotransposons, are widespread throughout eukaryote genomes. The socalled long interspersed elements-1 (LINE-1 or L1) are of especial interest because they represent the only class of retrotransposons in the human genome that are fully autonomous, meaning they encode all of the proteins required for their own retrotransposition. LINEs are transcribed into an RNA copy which is then reverse transcribed to create DNA that then integrates into the host genome, thereby expanding its copy number. Integration of these DNA copies can occur at a new site within the genome thereby creating a form of insertional mutagenesis. Thus, these elements are sometimes viewed as an important source of evolutionary drive; as in some cases, a new insertion can result in increased evolutionary fitness and provide the individual with an advantage, while in others it is detrimental [1,4]. LINEs make up approximately 17% of the human genome with an estimated half a million copies per genome. Retrotransposons in general constitute close to fifty percent of the human genome but are difficult to study in genomic or gene expression analyses. This is due to the highly-repeated nature of their sequences so that mapping these sequences to specific loci is challenging. Usually, therefore, in general genomic analyses, retrotransposons are deliberately masked or excluded. When human genome sequencing efforts first reported draft analyses of the genome, TEs were widely considered as junk that had accumulated over millions of years of evolution [5]. A number of recent observations stimulated interest in these elements. One was the discovery that retrotransposons were expressed in neurons of the adult brain [6,7]. This raised the prospect that insertional mutagenesis, caused by these elements, might affect neuronal function and in doing so increase neuronal plasticity. Although that latter idea is not universally accepted [8-10] there is still strong interest in understanding what role transposable elements play in neuronal function. Another observation that sparked renewed interest in retrotransposons was the observation that retrotransposons, which are transcriptionally silenced in most cell types of the body, can become re-activated in certain cancers [11]. These observations suggested that transcriptional activation of these elements, rather than just being a consequence of cellular transformation, could be the cause of transformation or, at the very least, contribute to disease progression. With an average size of 6 kb, LINEs encode two proteins from open reading frames (ORFs), a nucleic acid-binding protein often referred to as an RNA chaperone (ORF1p) and a protein that has endonuclease and reverse transcriptase activity (ORF2p). A primate-specific ORF transcribed in an antisense orientation, named ORF0, has recently been described although its’ function is still to be defined [12,13]. ORF1p and ORF2p are required for LINE-1 retrotransposition [14,15]. Interestingly, some other TEs, such as the shortinterspersed elements (SINEs), of which Alu elements are a part, are non-autonomous and require the LINE-1 proteins for their own retrotransposition [16-18]. This is important because SINE elements make up approximately 13% of the human genome and several SINEs (including Alus) are associated with human disease [reviewed in 1]. Therefore, the LINE elements and their encoded proteins can have major effects on the biology of our species. Because of this there is an important need to understand how LINE elements and their proteins function. Recently we isolated a protein complex associated with the ORF1p in human embryonic stem cells (hESCs), one of the few types of normal cells to naturally express LINE-1 elements [19]. Our studies build upon a number of groundbreaking studies that helped define the ORF1p-associated complex in other cells types [20-23]. Additionally, a recent genetic screen conducted in two different cell types identified genes involved in both inhibition and activation of LINE- 1 activity in mammalian cells [24]. The data from this genetic screen also highlight some of the proteins that likely restrict or activate LINE function in cells and data that is comparable to the proteomic studies. Finally, a recent study also examined the L1 elements (and especially ORF1p) during primate evolution [25] and provided new insights into how ORF1p might interact with cellular RNAs and proteins. Here we review the progress in our understanding of the L1-derived ORF1 protein and its interactors, how this information might inform our understanding of the potential role of LINE elements in human biology, and how proteomic strategies might be used to further these studies.
The ORF1p Complex in Cells
In early human pre-implantation development, LINEs are found to be expressed up to the blastocyst stage at which time the embryo is about to implant in the uterine wall [see 26]. At this time the embryo has two main cell types, an outer layer of trophectroderm cells that will give rise to part of the placenta, and an inner group of so-called epiblast cells that are pluripotent and can give rise to every cell type present in the body [see 27 for recent review]. In the next few days of development, LINEs are silenced in most somatic lineages but remain active in the germline [26]. In the germ cell lineage, unique mechanisms have evolved in order to control activity of TEs, including LINEs, since excessive TE-mediated insertional mutagenesis would likely have drastic effects on the genome. But most epiblast-derived cells that go on to form the somatic cells of the animal suppress the activity of transposable elements. Most likely, it is thought, that expression of TEs can lead to insertional mutagenesis with obvious prospects for creating deleterious effects on cells. In order to study the L1-derived ORF1p protein, a number of studies expressed ORF1p in cultured cells that do not normally express the protein as a way to identify potential ORF1p interactors [20-23]. These studies provided a number of important insights into how ORF1p might function. Building on these foundational studies, we isolated an ORF1p-associated complex from human embryonic stem cells (hESCs). These cells are derived directly from the pluripotent epiblast cells of the early human embryo in which LINE elements are naturally expressed. Indeed, hESCs mirror their in vivo counterparts in many ways, including in the natural expression of LINE elements [19]. Thus, we employed these cells because they naturally express TEs and notably express the LINE-derived protein ORF1p. Therefore, unlike the previous studies, there was no need to exogenously express ORF1p from various promoters. The overlap in proteins identified in our study and the previous studies was only about fifteen percent, suggesting that the composition of L1 ORF1p complex might be cell-type specific, affected by overexpression of the ORF1p in cells that don’t normally express it, affected by varying methods of isolating the ORF1p complex, or all of the above.
Here we will focus on two of the proteins identified in our screen [19] and many of the other screens [20-23], namely MOV10 and PURA, and discuss how recent studies have informed our views of how these proteins may impact ORF1p activity and, therefore, affect LINE-1 activity. The MOV10 helicase was also identified in a CRISPR–Cas9 genetic screening strategy in two distinct human cell lines [24]. In this study, the authors carried out a genome-wide survey of genes involved in the control of L1 retrotransposition. Amongst the genes identified in this study as suppressors of L1 activity was MOV10, consistent with previous studies that indicated that MOV10 acts as a restriction factor for retrotransposons and retroviruses [28-31]. Recent studies in C. elegans found that the worm homolog of MOV10, ERI- 6/7 acting together with Adenosine Deaminases Acting on RNAs (ADARs), is able to silence both long terminal repeat (LTR) retrotransposons and endogenous retroviruses in that system [32]. Interestingly, the authors of that study concluded that: “The activation of retrotransposons in the ADAR- and ERI-6/7/MOV10-defective mutant is associated with the induction of the unfolded protein response (UPR), a common response to viral infection” [32]. This work speculated that, since the proteins produced in response to viral infection and pathogens overlap with those expressed with retrotransposons, it might mean that there is a common response to “foreign elements” as has been suggested previously [see 33 for review]. Further the authors went on to suggest that this common response to foreign elements might occur because of the burden of having to replicate foreign RNA and proteins in the cell, a process that they termed: “proteotoxicity”. Interestingly, previous studies in HeLa cells demonstrated that ADAR1 can bind to the L1 RNP complex and inhibit retrotransposition [34]. It was also concluded that this inhibition of L1 retrotransposition was independent of ADAR1’s RNA editing function. Overall, the conclusion of this previous study was that ADAR1 didn’t interfere with the accumulation of the L1 RNP complex but rather somehow interfered with its’ activity [34]. Therefore, these new studies in C. elegans on the cooperativity of ADAR and ERI-6/7/MOV10 in retrotransposon silencing could point to new avenues of research into L1 element silencing in mammalian cells. Such studies could provide new insights into how MOV10 (and ADARs) might act in transposon silencing, findings that might impact our understanding of how mammalian cells could deal with a LINE-1-derived ORF1p complex. The notion that cells react to the burden of production of LINE-1-derived proteins and RNAs as a form of “proteotoxicity” is also interesting. This is especially the case in light of evidence that in certain circumstances riboncleoprotein (RNP) complexes, created when proteins associate and coat viralor retrotransposon-derived RNAs, are produced in cells of the brain and in human cancers. An interesting question is whether the metabolism of cells that deal with L1-derived RNA- and protein-induced proteotoxicity is altered and how that might, in turn, affect the stability of normal cell states?
Another recent study that shed light on the role of MOV10 focused on a group of emerging highly-pathogenic representatives of bunyaviruses [35]. These newly emerging viruses include severe fever with thrombocytopenia syndrome virus, considered by the World Health Organization to be a very dangerous pathogen [see 36 for brief review]. Bunyavirus RNP complexes can act both as a complex for RNA synthesis and as the structural core of the Bunyavirus virions. The RNPs are assembled when multiple copies of the Bunyavirus nucleoprotein coat the viral RNA and associate with a viral polymerase. This RNP complex thus represents the machinery for viral genome replication and transcription. The recent study showed that MOV10 associates with bunyavirus nucleoproteins and inhibits the replication of the virus, and thus restricts bunyaviral infection [35]. MOV10 does this by interfering with the nucleoprotein-arm domain (which consists of only 34 amino-acids), of nucleoprotein machinery and consequently disassembling the RNP complex [35]. This study therefore provides important new insights into how MOV10 might associate with the L1 ORF1p complex to restrict LINE-1 activity and perhaps also methods to develop inhibitors of ORF1p activity.
Another protein found in many studies of the ORF1p complex, including ours, is the purine rich element binding protein alpha (Pur-a) - a DNA- and RNAbinding protein encoded by the PURA gene [37,38]. PURA was also identified in the genome-wide CRISPRCas9 mediated screen described above as an inhibitor of L1 transposition [24]. In humans, the PURA gene has been identified as being involved in activating nuclear transcription, facilitating cytoplasmic RNA transport and regulating DNA replication in the cell cycle. Mutations in PURA have been associated with autosomal dominant and neurodevelopmental disorders [39]. Using genome sequencing methods, mutations in the PURA gene have been identified in patients with a range of neurological disorders including seizures, hypotonia, learning disabilities and developmental delay, now described as PURA syndrome [40]. As patients with mutations in the PURA gene continue to be identified, these findings expand our understanding of the functional domains of the PURA protein. Most recently a patient was described with a nonsense mutation within the PURA gene that expanded the range of phenotypes associated with PURA mutations to include short stature, problems with bone development and delayed puberty [36]. These studies do not immediately provide new information on how PURA might act at the molecular level or interact with L1-derived ORF1p. However, because both PURA and L1 ORF1p are expressed in the brain and PURA deficits affect brain development, these findings provide new avenues of exploration for understanding how LINE-1 retrotransposons might act in the brain during homeostasis and development. Intriguingly, mutations in both MOV10 and PURA are found to affect brain development [37,38,41,42]. One can therefore speculate on the role of ORF1p in the absence of, or mutations in, such proteins. Some key questions to consider: How does an ORF1p complex behave in the absence of Pur-α or MOV10? How do such conditions affect the transposition efficiency of LINE-1 and other L1- dependent elements, such as SINEs, in the brain? What impact does L1 ORF1p have on the functional activity of proteins like Pur-α and MOV10 in the brain of a normal individual? As the role of transposable elements in brain development and aging are examined more fully, it will be important to understand what role not only LINE-1- derived insertions play but also LINE-1-derived proteins, especially the most abundant, ORF1p.
New Roles for LINE-derived ORF1 Protein
While many of the recent studies described above pertain to the already known functions of ORF1p and its associated proteins, other recent studies suggest a new, and intriguing, aspect of ORF1p biology, namely in the relationship to newly-acquired viruses, including SARSCoV- 2. These studies described the ability of the SARSCoV- 2 virus, the cause of COVID-19 disease in humans, to integrate into the DNA of human cells in culture [43]. The ability of the SARS-CoV-2 to integrate into the human genome could, perhaps, create conditions for re-infection of an individual from a genomic pool. The ability of viruses such as SARS-CoV-2 to integrate into the genome would, presumably, require access to reverse transcriptase activity. This could, the authors suggest, be supplied by active LINE-1 elements. Indeed, the study found that cells in which there was greater evidence for insertion of SARSCoV- 2 into the genome had a much higher incidence of ORF1p expression than other cells. These data suggest that the ORF1p (presumably together with the L1-derived reverse transcriptase, ORF2p) could help novel viruses to enter the human genome. Importantly in this regard, the authors also showed that viral infection per se increases the expression of LINE-1 elements and the expression of ORF1p. One conclusion that can be drawn from these studies is that SARS-CoV-2 itself somehow stimulates the expression of LINE-1 elements, perhaps through some form of stress response (see above). In turn, the activated LINEs and their expressed products may help the virus integrate into the host. Others however have both questioned the methodology of the work [44] or found little evidence for SARS-CoV-2 integration into the genome of infected individuals [45] raising questions about the solidity of the other findings. Interestingly however, in terms of the ability of a virus to stimulate LINE-1 activity, previous studies examined the effect of the Kaposi’s sarcoma (KS)- associated herpesvirus (KSHV) on L1 retrotransposition [46,47]. KSHV is the causative agent of Kaposi’s sarcoma, a type of tumor commonly found in AIDS patients, as well as KSHV inflammatory cytokine syndrome, primary effusion lymphoma and HHV-8-associated multicentric Castleman’s disease and KSHV inflammatory cytokine syndrome. KSHV is one of several known “oncoviruses” or human cancer viruses. These studies indicated that KSHV infection led to down-regulation of MOV10 which in turn led to the up-regulation of L1 expression [46]. Therefore, it will be important to understand how the SARS-CoV-2 virus might, like KSHV, stimulate L1 expression, irrespective of whether it leads to SARS-CoV-2 integration into the genome.
Has Evolution of ORF1p Allowed Other Mobile Elements to Cross the Line?
One productive avenue of investigation into L1 ORF1p structure to function relationships has been to examine how the ORF1p itself evolves in multiple species as it has done in primates. Recently Furano, Jones and colleagues described an analysis of L1 ORF1p in different primate species [48]. The c-terminus of the ORF1p, which is highly conserved among L1 families, has multiple activities that are required for L1 retrotransposition. These activities include high affinity nucleic acid binding and chaperone activity, swift formation of stable nucleoprotein complexes and, of course, retrotransposition itself. The N-terminus of the ORF1p molecule contains a coiled-coil domain whose function is less clear but whose sequence has nevertheless altered over evolutionary time. These studies found that ORF1p can be susceptible to inactivation by single point mutations and, at the same time, can survive multiple amino acid substitutions [48]. According to Furano, Jones and colleagues, the evolution of the coiled-coil domains in mice and humans was proposed to be an adaptation of the ORF1p to genetic changes extrinsic to L1, that is, in its’ environment. Instead, they now argue that expansion of the coiled-coil domains occurs during evolution with some variability in the sequence of each domain. The authors suggest that by expanding the functional sequencing space of the coiled-coil domain, this mitigates any substitution events that could destroy ORF1p function, thus allowing L1 to thrive over many evolutionary events. Perhaps by expanding the coiled-coil domain, L1 elements made themselves more adaptable and more resistant to damaging mutations that would kill their retrotransposition. An intriguing idea, but does L1-derived ORF1p adaptability come at a cost to its host?
It has been known for some time that L1-derived proteins are required for the retrotransposition of other elements including SINEs and Alus, a subset of SINEs [see 49 for review]. More recently it has become apparent that L1- derived proteins are required for the transposition of other non-autonomous elements. Some of these elements include so-called composite elements (named after their constituent elements) and include variable number of tandem repeats (VNTRs) including SVA (SINE-R-VNTRAlu) and LAVA (L1-Alu-VNTR-Alu). Recent studies examined the ability of the L1 ORF1p to mobilize a hominid-specific SVA element and found that this element is a preferred substrate of the L1-derived proteins [25]. These studies also confirmed, using in vitro assays, that the SVA-D element is an active, albeit non-autonomous, human transposable element. The implications of these findings are that L1-derived ORF1p can aid in the transposition of the SVA element. Overall, this study was not able to conclude that L1-derived ORF1p co-evolved with composite elements. It did nevertheless find that changes in ORF1p in Orangutans, for example, were associated with a reduced genomic insertion rate of SVA retrotransposons in that species by comparison with that seen in humans. A speculation from this latter finding is that changes in the Orangutan ORF1p coding sequence made SVA transposons a less favored co-factor for SVA RNA. An alternative way of looking at this is that the human L1 still adequately supports SVA retrotransposition. Why should we care about SVA retrotransposition since previous studies have suggested that the retrotransposition rate of SVA within the human genome is fairly small? One reason to be interested is that more recent analyses of SVA retrotranspostion rates suggest a higher rate of SVA insertions in the human genome than had previously been thought [50]. This study analyzed de novo L1, SVA, and Alu retrotransposition events in 599 individuals in the Utah Centre d’Etude du Polymorphisme Humain (CEPH) study, comprising 33 three-generation pedigrees. Analysis of insertion events in these individuals was carried out using three mobile element insertion-software calling tools. The insertion rate for Alu elements was estimated to be one in 40 births, which was lower than previous estimates. The insertion rate for L1s was estimated to be 1 in 63 births and was within the range of the previous estimates which ranged from 1 in 20 to 1 in 200 births. But when the authors estimated the rate of SVA insertions within these pedigrees they found that the insertion rate was 1 in 63. This was much higher than previous estimates which had suggested an SVA insertion rate of 1 in 900 births. Obviously, new insertions such as these could have both beneficial and harmful effects on the genome and the individual. Nevertheless, perhaps this is a cause for concern and a reason to try and better understand how the L1 ORF1p facilitates transposition of SVA and other elements and what impact environmental factors might have on L1 activation during embryonic and fetal development?
Some of these recent findings about the ORF1p of LINE- 1 elements indicate that it could have a much broader impact on human biology than simply enabling the retrotransposition of its’ own element. All in all, they suggest that the ORF1p can bind not only its’ own RNA but also the RNA of other retrotransposons and perhaps even of viruses. Another recent study examined the ability of the L1 ORF1p to bind other cellular RNAs. Brigg, Kerrow and colleagues examined the ORF1p complex in prostate cancer cells which express ORF1p [51]. Cancer cells, unlike their normal counterparts, often express retrotransposons [reviewed in 2]. Indeed, ORF1p is found to be expressed in approximately fifty percent of all tumors, and these tumors are also found to have some L1 insertions [2,11]. By immunoprecipitating the ORF1p complex from prostate cancer cells and examining associated RNAs, these authors found that not only did the ORF1p bind to its’ own RNA but also to normal cellular RNAs including circular RNAs, poly-A+ RNA and processing-body (P-body)-associated RNA. Indeed, they found that LINE-1 RNA was only a small component of the ORF1p-associated complex and that other RNAs actually were in the vast majority [51]. An interesting finding of this study was that the ORF1p complex-associated or P-body-associated RNAs were correlated with L1 RNA expression in prostate cancer cell lines. Using a knockdown approach to reduce ORF1p levels, the authors observed a down-regulation of RNAs associated with P-bodies. Because P-bodies have been associated with controlling a variety of key processes including RNA processing, protein synthesis, protein degradation and chromatin regulation [see 52, 53 for reviews], ORF1p could, conceivably, interfere with any of these processes. The authors noted: “In particular, almost all of the genes that are significantly correlated with LINE- 1 RNA in prostate cancer were also enriched in processing bodies (P-bodies)” [51]. P-bodies are distinct foci formed in cells through the process of phase separation and contain many enzymes responsible for RNA turnover. Therefore, these recent studies indicate that LINE-1 may interfere with the degradation of P-body-associated RNAs [51]. Again, the consequence of ORF1p interfering in RNA processing or degradation in cells could be dramatic. Intriguingly, P-body homeostasis plays a critical role in the maintenance of the pluripotent state in mouse embryonic stem cells [54]. Thus L1-derived impairment of P-body RNA processing in somatic cells (perhaps even somatic stem cells) could conceivably alter their state of differentiation with, perhaps, consequences for their ultimate malignant transformation.
Conclusion
Despite the fact that L1 elements make up about 17% of the human genome, until relatively recently they were considered junk, relics of our ancient past. Proteomic studies have elucidated some of the proteins that interact with those produced by L1 elements, especially ORF1p. Although the impact of L1 element-derived insertions on the human genome is still being assessed, because every time these elements are expressed they produce ORF1p, it is becoming clear that this protein could have a major role in our biology. Recent proteomic, genetic and biochemical studies have increased our understanding of how ORF1p interacts with cellular (and viral) RNAs and proteins and provides a much more complex picture of its’ potential role in cellular physiology (Figure 1). Altogether, the recent findings highlighted here have implications for why it might be interesting to target L1 ORF1p for inhibition in humans, a strategy that might be useful for treating a number of conditions in which the ORF1p is found to be expressed, especially cancer. In that regard the genomewide screen described recently might be one place to look for targets though which to target ORF1p activity because that screen identified suppressors of L1 activity in addition to activators [24]. It will also be interesting to see if and how divergent L1-derived ORF1p molecules found in primate species could alter the substrate-specificity of ORF1p for exogenous viruses and, therefore, alter the host range of such infectious agents. All in all, we may need to pay much more attention to the so-called “Dark Side” of the genome and the proteins it produces. Proteomic studies that expand upon previous studies could be very useful in determining whether the differences observed in proteins associated with ORF1p are method-specific or cell type-specific. Moreover, new methods of isolating associated proteins with improved proximity tags for mass spectroscopy could provide new data on direct versus indirect interactions between ORF1p and components of the ribonucleoprotein complex. Similarly, proteomic analyses of post-translational modifications of the L1 RNP complex and its’ associated proteins might help elucidate potential mechanisms by which the cell deals with such a complex. Understanding how L1 elements and their encoded proteins function, both in their own life cycle and in the life cycle of other transposable elements and viruses, could provide new insights into the impact that all mobile elements play in the human life cycle, in human health and on the human genome.
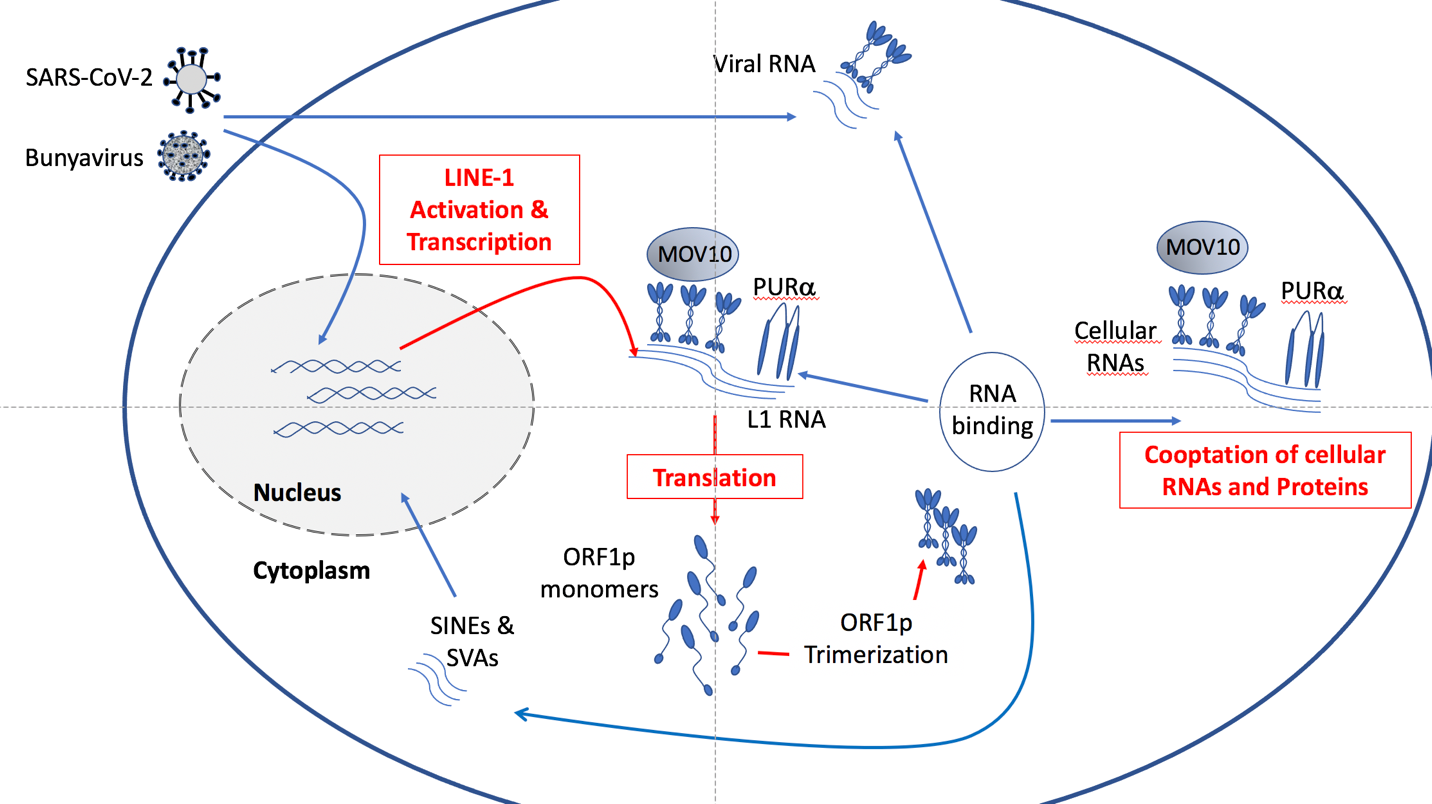
Figure 1: L1 ORF1p interactions in cells as defined by proteomic analysis. Activation of L1 elements by whatever means leads to the production of L1 RNA that is then transcribed into ORF2 and ORF0 (not shown) and ORF1p. ORF1p monomers form trimers that can then bind the L1 RNA and enable the L1 elements to transpose. Production of L1 RNA and proteins could place new metabolic needs on the cell producing them (highlighted in red). In addition, the ORF1 protein can likely bind both normal cellular RNAs and proteins, perhaps interfering with their normal function. This could create a state of “proteotoxicity” that might exacerbate other stresses on the cell, perhaps leading to cell death or engulfment by the immune system. Activation of L1 elements by viruses could provide a mechanism by which they use the L1 ORF1P to enable their own life cycles. The consequences of this latter action remain to be fully determined. Finally, other transposable elements such as SINEs (including ALUs) and SVA use the L1-machinery to transpose, potentially creating new insertions in the human genome.
Acknowledgements
We thank our colleagues Fabio Macciardi and Suzanne Sandmeyer for helpful discussions and comments on the manuscript. We apologize in advance to any colleagues in the field whose work we did not cite because of space constraints or our own ignorance.
References
2. Burns KH. Transposable elements in cancer. Nat Rev Cancer. 2017 Jul;17(7):415-424.
3. Suarez NA, Macia A, Muotri AR. LINE-1 retrotransposons in healthy and diseased human brain. Dev Neurobiol. 2018 May;78(5):434-455.
4. Gorbunova V, Seluanov A, Mita P, McKerrow W, Fenyö D, Boeke JD, et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature. 2021 Aug;596(7870):43-53.
5. Dubnau J. The Retrotransposon storm and the dangers of a Collyer’s genome. Curr Opin Genet Dev. 2018 Apr;49:95-105.
6. Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005 Jun 16;435(7044):903-10.
7. Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009 Aug 27;460(7259):1127-31.
8. Evrony GD, Lee E, Park PJ, Walsh CA. Resolving rates of mutation in the brain using single-neuron genomics. Elife. 2016 Feb 22;5:e12966.
9. Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012 Oct 26;151(3):483-96.
10. Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011 Oct 30;479(7374):534-7.
11. Rodic N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol. 2014 May;184(5):1280-6.
12. Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MC, et al. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell. 2015 Oct 22;163(3):583-93.
13. Honda T, Nishikawa Y, Nishimura K, Teng D, Takemoto K, Ueda K. Effects of activation of the LINE-1 antisense promoter on the growth of cultured cells. Sci Rep. 2020 Dec 17;10(1):22136.
14. Martin SL, Cruceanu M, Branciforte D, Wai-Lun Li P, Kwok SC, Hodges RS, et al. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J Mol Biol. 2005 May 6;348(3):549-61.
15. Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE- 1 retrotransposition. Hum Mol Genet. 2005 Nov 1;14(21):3237-48.
16. Dewannieux M, Esnault C, Heidmann T. LINEmediated retrotransposition of marked Alu sequences. Nat Genet. 2003 Sep;35(1):41-8.
17. Dewannieux M, Heidmann T. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J Mol Biol. 2005 Jun 3;349(2):241-7.
18. Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, et al. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2012 Feb;40(4):1666-83.
19. Vuong LM, Pan S, Donovan PJ. Proteome Profile of Endogenous Retrotransposon-Associated Complexes in Human Embryonic Stem Cells. Proteomics. 2019 Aug;19(15):e1900169.
20. Taylor MS, LaCava J, Mita P, Molloy KR, Huang CR, Li D, et al. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell. 2013 Nov 21;155(5):1034-48.
21. Taylor MS, Altukhov I, Molloy KR, Mita P, Jiang H, Adney EM, et al. Dissection of affinity captured LINE-1 macromolecular complexes. Elife. 2018 Jan 8;7:e30094.
22. Moldovan JB, Moran JV. The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition. PLoS Genet. 2015 May 7;11(5):e1005121.
23. Goodier JL, Cheung LE, Kazazian HH Jr. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 2013 Aug;41(15):7401-19.
24. Liu N, Lee CH, Swigut T, Grow E, Gu B, Bassik MC, Wysocka J. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature. 2018 Jan 11;553(7687):228-232.
25. Damert A. LINE-1 ORF1p does not determine substrate preference for human/orangutan SVA and gibbon LAVA. Mob DNA. 2020 Jul 11;11:27.
26. Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, et al. The DNA methylation landscape of human early embryos. Nature. 2014 Jul 31;511(7511):606-10.
27. Zhai J, Xiao Z, Wang Y, Wang H. Human embryonic development: from peri-implantation to gastrulation. Trends Cell Biol. 2021 Aug 17:S0962-8924(21)00148-3.
28. Peddigari S, Li PW, Rabe JL, Martin SL. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res. 2013 Jan 7;41(1):575-85.
29. Arjan-Odedra S, Swanson CM, Sherer NM, Wolinsky SM, Malim MH. Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology. 2012 Jun 22;9:53.
30. Li X, Zhang J, Jia R, Cheng V, Xu X, Qiao W, et al. The MOV10 helicase inhibits LINE-1 mobility. J Biol Chem. 2013 Jul 19;288(29):21148-21160.
31. Goodier JL, Cheung LE, Kazazian HH Jr. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012;8(10):e1002941.
32. Fischer SEJ, Ruvkun G. Caenorhabditis elegans ADAR editing and the ERI-6/7/MOV10 RNAi pathway silence endogenous viral elements and LTR retrotransposons. Proc Natl Acad Sci U S A. 2020 Mar 17;117(11):5987-5996.
33. Gazquez-Gutierrez A, Witteveldt J, R Heras S, Macias S. Sensing of transposable elements by the antiviral innate immune system. RNA. 2021 Apr 22;27(7):735-52.
34. Orecchini E, Doria M, Antonioni A, Galardi S, Ciafrè SA, Frassinelli L, et al. ADAR1 restricts LINE- 1 retrotransposition. Nucleic Acids Res. 2017 Jan 9;45(1):155-168.
35. Mo Q, Xu Z, Deng F, Wang H, Ning YJ. Host restriction of emerging high-pathogenic bunyaviruses via MOV10 by targeting viral nucleoprotein and blocking ribonucleoprotein assembly. PLoS Pathog. 2020 Dec 7;16(12):e1009129.
36. Windhaber S, Xin Q, Lozach PY. Orthobunyaviruses: From Virus Binding to Penetration into Mammalian Host Cells. Viruses. 2021 May 10;13(5):872.
37. Daniel DC, Johnson EM. PURA, the gene encoding Pur-alpha, member of an ancient nucleic acid-binding protein family with mammalian neurological functions. Gene. 2018 Feb 15;643:133-143.
38. Molitor L, Bacher S, Burczyk S, Niessing D. The Molecular Function of PURA and Its Implications in Neurological Diseases. Front Genet. 2021 Mar 11;12:638217.
39. Reijnders MRF, Leventer RJ, Lee BH, Baralle D, Selber P, Paciorkowski AR, et al, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2021.
40. Cinquina V, Ciaccio C, Venturini M, Masson R, Ritelli M, Colombi M. Expanding the PURA syndrome phenotype: A child with the recurrent PURA p.(Phe233del) pathogenic variant showing similarities with cutis laxa. Mol Genet Genomic Med. 2021 Jan;9(1):e1562.
41. Skariah G, Seimetz J, Norsworthy M, Lannom MC, Kenny PJ, Elrakhawy M, et al. Mov10 suppresses retroelements and regulates neuronal development and function in the developing brain. BMC Biol. 2017 Jun 29;15(1):54.
42. Skariah G, Perry KJ, Drnevich J, Henry JJ, Ceman S. RNA helicase Mov10 is essential for gastrulation and central nervous system development. Dev Dyn. 2018 Apr;247(4):660-671.
43. Zhang L, Richards A, Barrasa MI, Hughes SH, Young RA, Jaenisch R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc Natl Acad Sci U S A. 2021 May 25;118(21):e2105968118.
44. Parry R, Gifford RJ, Lytras S, Ray SC, Coin LJM. No evidence of SARS-CoV-2 reverse transcription and integration as the origin of chimeric transcripts in patient tissues. Proc Natl Acad Sci U S A. 2021 Aug 17;118(33):e2109066118.
45. Chen YS, Lu S, Zhang B, Du T, Li WJ, Lei M, et al. Comprehensive analysis of RNA-seq and whole genome sequencing data reveals no evidence for SARS-CoV-2 integrating into host genome. Protein Cell. 2021 Aug 2:1-7.
46. Nakayama R, Ueno Y, Ueda K, Honda T. Latent infection with Kaposi’s sarcoma-associated herpesvirus enhances retrotransposition of long interspersed element-1. Oncogene. 2019 May;38(22):4340-4351.
47. Ahuja A, Journo G, Eitan R, Rubin E, Shamay M. High levels of LINE-1 transposable elements expressed in Kaposi’s sarcoma-associated herpesvirusrelated primary effusion lymphoma. Oncogene. 2021 Jan;40(3):536-550.
48. Furano AV, Jones CE, Periwal V, Callahan KE, Walser JC, Cook PR. Cryptic genetic variation enhances primate L1 retrotransposon survival by enlarging the functional coiled coil sequence space of ORF1p. PLoS Genet. 2020 Aug 14;16(8):e1008991.
49. Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV. The Influence of LINE- 1 and SINE Retrotransposons on Mammalian Genomes. Microbiol Spectr. 2015 Apr;3(2):MDNA3-0061-2014.
50. Feusier J, Watkins WS, Thomas J, Farrell A, Witherspoon DJ, Baird L, et al. Pedigree-based estimation of human mobile element retrotransposition rates. Genome Res. 2019 Oct;29(10):1567-1577.
51. Briggs EM, McKerrow W, Mita P, Boeke JD, Logan SK, Fenyö D. RIP-seq reveals LINE-1 ORF1p association with p-body enriched mRNAs. Mob DNA. 2021 Feb 9;12(1):5.
52. Luo Y, Na Z, Slavoff SA. P-Bodies: Composition, Properties, and Functions. Biochemistry. 2018 May 1;57(17):2424-2431.
53. Standart N, Weil D. P-Bodies: Cytosolic Droplets for Coordinated mRNA Storage. Trends Genet. 2018 Aug;34(8):612-626.
54. Pecori F, Kondo N, Ogura C, Miura T, Kume M, Minamijima Y, et al. Site-specific O-GlcNAcylation of Psme3 maintains mouse stem cell pluripotency by impairing P-body homeostasis. Cell Rep. 2021 Jul 13;36(2):109361.