Abstract
In accordance with the present epidemiological paradigm, viral mutations of the virus are on the rise, and their natural effects are being selected for at a higher rate than normal. According to the World Health Organization (WHO), the global COVID-19 pandemic induced by the Delta and Omicron strain of the SARS-CoV-2 virus could propagate and disseminate more rapidly than other viruses thanks to its many mutations, and these also caused some very significant health problems. The established medications would eventually start to lose their efficacy since the variation mutated more quickly than the original stain. As protein spikes are the point of origin or epitome for the mutations to take place, it would be most effective to target the remaining vital enzymes by binding the proteins with the largest pocket sizes. The objective of the current work is to employ in-silico analysis to discover the streptomyces chemicals that suppress the SARS-CoV-2 virus as well as its mutated strains thus promoting a healthy body. Based on the drug likeness property of compounds when subjected to molecular docking, a total of 14 compounds were identified and selected from the PUBCHEM database that showed highest binding energy with the targeted Receptor Binding Domain. The compounds namely - Streptomyces tanashiensis; Thaxtomin A; Bafilomycin A1 from Streptomyces griseus and few others as mentioned further on more research would support and confirm the utilizing of these to create new medications to treat the novel SARS-CoV-2 infectious strains.
Keywords
SARS-CoV-2, Saccharomyces, Streptomyces, Drug development, Ligands, Receptor binding domain (RBD), Drug likeness, Molecular docking
Introduction
Scientists have anticipated that Coronavirus is going to be endemic, albeit with diminished virulence and fatality rate, and will continue to produce variants. Omicron, Alpha, and Delta are some of the prominent variants. Few of these variants also tend to produce some likely sub-variants into nature. When a virus is transmitted from one individual to another, it sometimes faces some changes in the structure which in medical layman terms is also known as mutations. When two or more mutations occur, a variant is born. There were over 5000+ mutations discovered in 200,000 genome isolates present in the spike proteins of SARS-CoV-2.
Covid variants have the potential to emerge from animals, humans, and from infected animals and humans. We have seen its effect in humans but not much research has been in animals. Some variants may not be strong enough to be treated and hence can be cured easily over the time but some variants may be produced in the future which might have potential to bring pandemic once again. Some reports also claim that some of the variants and sub-variants are getting drug resistant.
According to WHO reports, as of February 2023, the confirmed cases of COVID-19 are a total of 756,411,740 and confirmed deaths are 6,842,462. Because of the restrictions implemented due to COVID-19 virus, it is challenging for humans to exist under them. So, there is a need for compounds which can be developed as drugs. It is the need of an hour to derive compounds from various different sources such as plants, chemicals, gasses, etc.
In this research, we have selected Streptomyces to derive potential compounds for further drug development because Streptomyces bacteria produce a significant portion of antibiotics used today [1]. Actinobactriae’s largest genus is Streptomyces [4]. A gram- positive bacteria’s genus i.e. Streptomyces grows in a variety of environments and resembles filamentous fungi in appearance [5]. A necessary component of Streptomyces’ morphological differentiation is the production layer of the hyphae that can differentiate into a chain of spores. The potential of Streptomyces to produce bioactive secondary metabolites, such as not only antibiotics but also antifungals, antivirals, antitumorales, antihypertensive-ness, and immunosuppressants, is particularly noteworthy. As the production of the majority of the antibiotics is species-specific, these secondary metabolites are Streptomyces species in order to compete with other bacteria that come into contact, even ones belonging to the same genre.
Around two-thirds of clinically relevant natural antibiotics, such as neomycin, cypemycin, grisemycin, bottromycin, and chloramphenicol, are produced by streptomyces [6,7]. From the genus Streptomyces comes the antibiotic streptomycin [8].
Streptomyces has 679 recognized species and an additional 121 tentative species. Only one species is used in development of COVID-19 drugs i.e. Streptomyces lividans. To find out more compounds that have potential for drug development, we have selected Streptomyces for our research due to its properties.
In-silico analysis is used for determining potential compounds because it is a computational method that uses computer simulation, modeling, and data analysis to investigate many areas of biology, chemistry, and other specific domains. It includes evaluating and interpreting data via the use of software programs and mathematical algorithms, as opposed to doing laboratory tests. In the context of drug development, in- silico analysis is utilized to anticipate the behavior of molecules and their interactions with target receptors or enzymes, in addition to optimizing drug design and identifying valuable therapeutic candidates. This makes this technique potential enough to start the research. This technique also has a number of benefits over conventional experimental approaches, including cost effectiveness, speed, and a thorough knowledge of complex biological systems.
Materials and Methods
Selection and preparation of ligands
Compounds produced in regards to Streptomyces species were identified and were about 190 in number. With the help of the official PUBCHEM website (https://pubchem/ncbi/nlm.nih.gov), the canonical smiles of the produced ligands were recovered. Further with the help of CORINA online server (https://demos.mn- am.com/corina.html), after embracing the SMILES string in the appropriate places the 3-D structure of each and every ligand was obtained for further steps. In the subsequent steps, by detecting the torsion root and correcting the angles of torsion, optimizing and assigning charges using the UFF i.e Universal Force Field, in order to make it easier to create the molecules’ 3D atomic coordinates, the ligands were created and transformed into PDBQT (Protein Data Bank, Partial Charge (Q), & the Atom type (T)) format.
Screening of the ligands based on druglikeness
Using the SwissADME online server (http://swissadme.ch/index.php), compounds’ drug-likeness were evaluated. The drug-likeness is an essential criterion for validating compounds as prospective ligands against potential therapeutic targets. Using Lipinski's Rule of Five, the 190 identified Streptomyces phytochemical compounds were screened and the positive drug likeness showing compounds were further used for docking studies.
Preparation of target proteins and identification of binding sites
The RBD of SARS-CoV-2, which is shown in Figure 1, served as the study's primary focus. Using the Protein data bank reserves (http://www.rcsb.org/), the 3D crystallographic structure of the proteins was retrieved. Using the PyMol software server (http://www.pymol.org) the proteins were visualized and the ligands, water molecules and co-crystal ligand structures which were bound to the COX-2 were further removed while processing. By using an open-source software Auto Dock Tools, the protein was prepared by adding charges and energy minimization in the Swiss PDB viewer by converting it subsequently into PDBQT format. Further with the help of CASTp online server, the binding sites for every protein were identified. The ligands having the largest pocket size of the protein were to be used as binding sites.
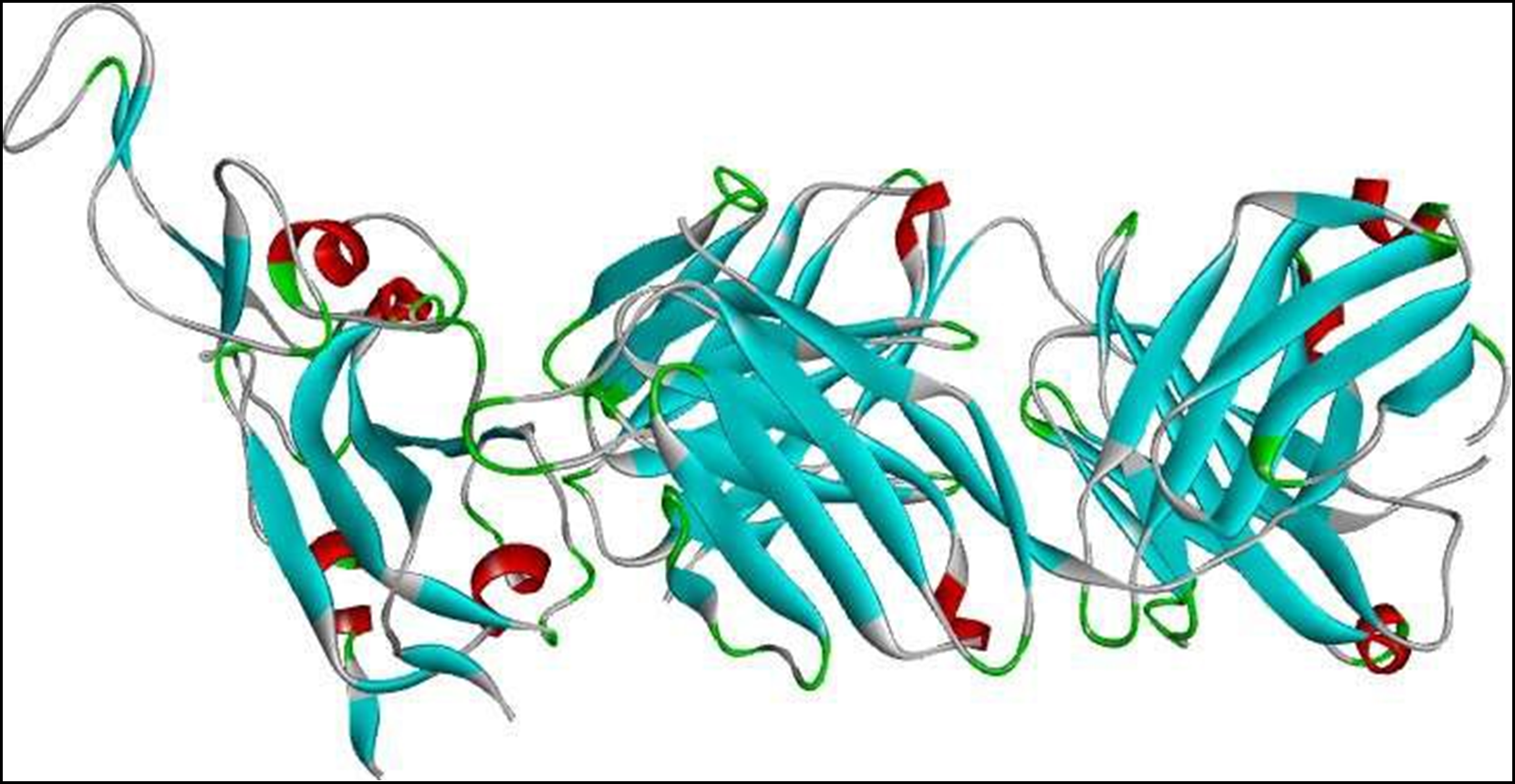
Figure 1. 3D structure of SARS-CoV-2 receptor binding domain (RBD).
Molecular docking and protein-ligand interactions analysis
The docking engine used for the molecular docking analysis was the PyRx tool from Auto Dock wizard. The protein was anticipated to be rigid during docking, but the ligands were thought to be flexible. Based on the binding amino acid analysis from CASTp server, they served as the foundation for the configuration file for the grid settings. The ligand with the greatest binding affinity i.e., the one with the most negative value was identified as having the highest binding energy. Using Biovia Drug discovery studio 2020, visual inspection of the docking site and Protein-Ligand interactions was carried out.
Results
Drug likeness analysis
From the drug likeness analysis of 190 compounds, 61 compounds showed drug likeness properties. Therefore 61 compounds were further subjected to docking and interaction analysis. Table 1 contains all the names of these compounds.
|
S.No |
Compound name |
|---|---|
|
1 |
Staurosporine, from Streptomyces species |
|
2 |
Manumycin A, Streptomyces parvulus |
|
3 |
Staurosporine, Streptomyces sp. |
|
4 |
InSolutionTM Leptomycin A, Streptomyces sp. |
|
5 |
From Streptomyces tanashiensis |
|
6 |
Thaxtomin A |
|
7 |
Glucose isomerase from streptomyces rubiginosus |
|
8 |
Bafilomycin A1 from Streptomyces griseus |
|
9 |
Platensimycin, >=90% (HPLC), from Streptomyces platensis |
|
10 |
Antibiotic from Streptomyces Sparsogenes |
|
11 |
Potassium clavulanate, from Streptomyces clavuligerus |
|
12 |
InSolutionTM Leptomycin B, Streptomyces sp. |
|
13 |
Streptomyces A-Factor |
|
14 |
Bafilomycin A1, Streptomyces griseus |
|
15 |
RK-682, Streptomyces sp. |
|
16 |
Mitomycin |
|
17 |
Trichostatin A |
|
18 |
Anisomycin |
|
19 |
Ionomycin calcium salt |
|
20 |
Leptomycin B |
|
21 |
Ionomycin |
|
22 |
Borrelidin |
|
23 |
2-Isocapryloyl-3R-hydroxymethyl-gamma-butyrolactone |
|
24 |
Ionomycin calcium |
|
25 |
Indanomycin |
|
26 |
6,7,12,13-Tetrahydro-5H-indolo[2,3-a] pyrrolo[3,4-c] carbazol-5-one |
|
27 |
8-Ethenyl-1-hydroxy-10,12-dimethoxy-4-[(2S,3S,4R,5S,6R)-3,4,5-trihydroxy-4,6-dimethyloxan-2-yl]naphtho[1,2-c]isochromen-6-one |
|
28 |
Azotomycin |
|
29 |
Aureothricin |
|
30 |
Lavendustin A |
|
31 |
A-Factor |
|
32 |
Toyocamycin |
|
33 |
Kendomycin |
|
34 |
Macromomycin B |
|
35 |
Tubercidin |
|
36 |
Cycloheximide |
|
37 |
Cerulomycin |
|
38 |
Antibiotic DC 107 |
|
39 |
Adenosine |
|
40 |
Mitomycin derivative T 41 |
|
41 |
Pyrroxamycin |
|
42 |
Thaxtomin A |
|
43 |
Acetoxycycloheximide |
|
44 |
Sparsomysin |
|
45 |
7-(4-(Dimethylamino)phenyl)-N-hydroxy-4,6-imethyl-7-oxo-2,4-heptadienamide |
|
46 |
Echinosporin |
|
47 |
Acetyloxycycloheximide |
|
48 |
Lactoquinomycin A |
|
49 |
Mitomycin C methylamine |
|
50 |
Leinamycin |
|
51 |
Leptomycin A |
|
52 |
Bafilomycin A1 |
|
53 |
N-(3-Hydroxy-1-oxocyclopent-2-en-2-yl)-3-(4-hydroxy-3-methoxyphenyl)propenamide |
|
54 |
(2S,3S,4S,6R)-3-Methoxy-2-methyl-4-(methylamino)-29-oxa- 1,7,17-triazaoctacyclo[12.12.2.12,6.07,28.08,13.015,19.020,27.021,26]nonacosa-8,10,12,14,19,21,23,25,27-nonaen-16-one |
|
55 |
CID 53230011 |
|
56 |
Azalomycin |
|
57 |
Schembl20768056 |
|
58 |
(3Z,5Z)-3,5-Bis(3,4-dichlorobenzylidene)-1-(3-morpholinopropanoyl)-4-piperidone;hydrochloride |
|
59 |
2-Amino-3-phosphino-propanoic acid |
|
60 |
Sparsomycin |
|
61 |
Triacsin c |
CastP Binding site analysis
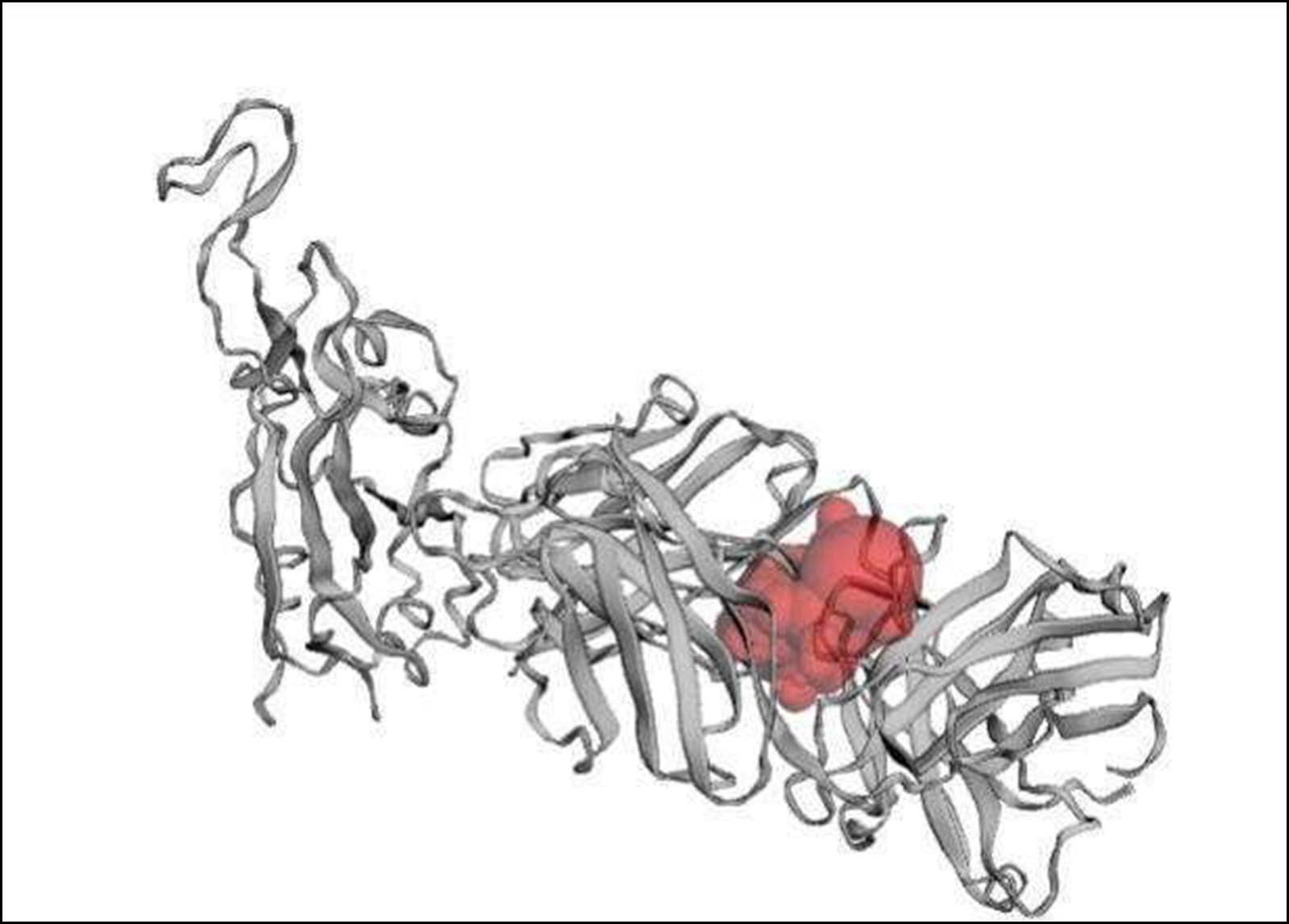
Figure 2. Binding pocket of the receptor binding domain (RBD).
|
Target Protein |
Amino acid residues in binding sites |
|---|---|
|
Receptor Binding domain |
H: 9-THR, 11-VAL, 39-GLN, 40-MET, 41-PRO, 42-GLY, 43-LYS, 87-THR, 88-ALA, 89-ILE, 91-TYR, 108-THR, 109-VAL, 110-THR, 112-SER, 146-PHE, 147-PRO, 148-GLU, 149-PRO, 167-PRO, 168-ALA, 169-VAL, 170-LEU, 174-GLY, 176-TYR L: 38-GLN, 39-LYS, 40-PRO, 41-GLY, 42-GLN, 83-VAL, 85-VAL, 87-TYR, 103-LYS, 104-VAL, 105-GLU, 142-ARG, 161-GLU, 162-SER, 163-VAL, 164-THR, 165-GLU, 166-GLN, 173-THR |
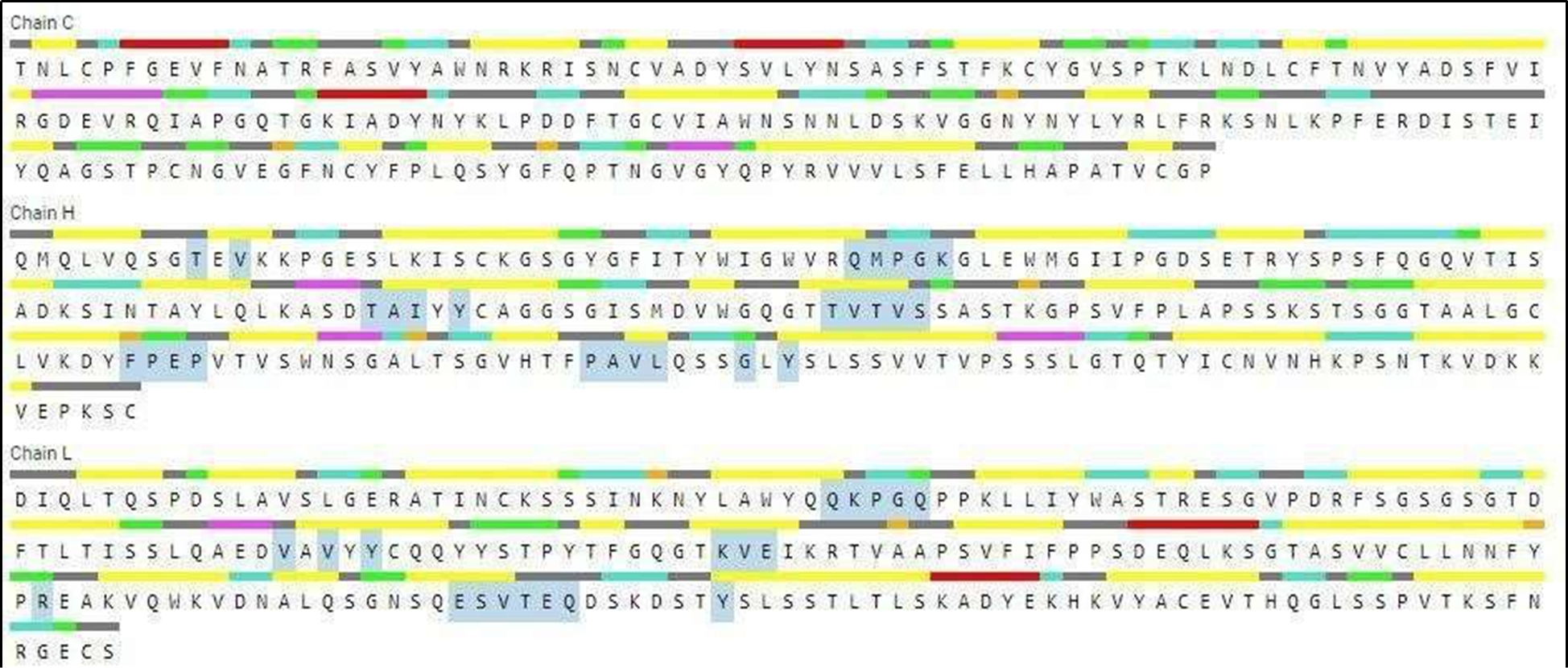
Figure 3. Aminoacid residues present involved in binding sites (highlighted in gray color).
Docking analysis
Table 3 depicts the binding energies of all the 61 compounds.
|
S.No |
Compound name |
Binding energy (Kcal/mol) |
|---|---|---|
|
1 |
Staurosporine, from Streptomyces species |
-7.6 |
|
2 |
Manumycin A, Streptomyces parvulus |
-7 |
|
3 |
Staurosporine, Streptomyces sp. |
-7.5 |
|
4 |
InSolutionTM Leptomycin A, Streptomyces sp. |
-6.1 |
|
5 |
From Streptomyces tanashiensis |
-8.9 |
|
6 |
Thaxtomin A |
-8 |
|
7 |
Glucose isomerase from streptomyces rubiginosus |
-7.4 |
|
8 |
Bafilomycin A1 from Streptomyces griseus |
-8.7 |
|
9 |
Platensimycin, >=90% (HPLC), from Streptomyces platensis |
-7.2 |
|
10 |
Antibiotic from Streptomyces Sparsogenes |
-6 |
|
11 |
Potassium clavulanate, from Streptomyces clavuligerus |
-6.1 |
|
12 |
InSolutionTM Leptomycin B, Streptomyces sp. |
-6.7 |
|
13 |
Streptomyces A-Factor |
-5 |
|
14 |
Bafilomycin A1, Streptomyces griseus |
-8.6 |
|
15 |
RK-682, Streptomyces sp. |
-5.3 |
|
16 |
Mitomycin |
-7.1 |
|
17 |
Trichostatin A |
-6.6 |
|
18 |
Anisomycin |
-5.9 |
|
19 |
Ionomycin calcium salt |
-6.5 |
|
20 |
Leptomycin B |
-6.5 |
|
21 |
Ionomycin |
-6.2 |
|
22 |
Borrelidin |
-8.2 |
|
23 |
2-Isocapryloyl-3R-hydroxymethyl-gamma-butyrolactone |
-4.7 |
|
24 |
Ionomycin calcium |
-6.4 |
|
25 |
Indanomycin |
-7.2 |
|
26 |
6,7,12,13-Tetrahydro-5H-indolo[2,3-a] pyrrolo[3,4-c] carbazol-5-one |
-8.4 |
|
27 |
8-Ethenyl-1-hydroxy-10,12-dimethoxy-4-[(2S,3S,4R,5S,6R)-3,4,5-trihydroxy-4,6-dimethyloxan-2-yl]naphtho[1,2-c]isochromen-6-one |
-7.8 |
|
28 |
Azotomycin |
-6.1 |
|
29 |
Aureothricin |
-5.2 |
|
30 |
Lavendustin A |
-6.8 |
|
31 |
A-Factor |
-5.2 |
|
32 |
Toyocamycin |
-6.4 |
|
33 |
Kendomycin |
-9.3 |
|
34 |
Macromomycin B |
-6.5 |
|
35 |
Tubercidin |
-6.5 |
|
36 |
Cycloheximide |
-6.3 |
|
37 |
Cerulomycin |
-5.8 |
|
38 |
Antibiotic DC 107 |
-8.5 |
|
39 |
Adenosine |
-6.7 |
|
40 |
Mitomycin derivative T 41 |
-6.5 |
|
41 |
Pyrroxamycin |
-6.9 |
|
42 |
Thaxtomin A |
-8 |
|
43 |
Acetoxycycloheximide |
-7.1 |
|
44 |
Sparsomysin |
-6.3 |
|
45 |
7-(4-(Dimethylamino)phenyl)-N-hydroxy-4,6-imethyl-7-oxo-2,4-heptadienamide |
-6.6 |
|
46 |
Echinosporin |
-6.5 |
|
47 |
Acetyloxycycloheximide |
-7.1 |
|
48 |
Lactoquinomycin A |
-8.9 |
|
49 |
Mitomycin C methylamine |
-6.2 |
|
50 |
Leinamycin |
-8.5 |
|
51 |
Leptomycin A |
-6.6 |
|
52 |
Bafilomycin A1 |
-8.5 |
|
53 |
N-(3-Hydroxy-1-oxocyclopent-2-en-2-yl)-3-(4-hydroxy-3-methoxyphenyl)propenamide |
-6.8 |
|
54 |
(2S,3S,4S,6R)-3-Methoxy-2-methyl-4-(methylamino)-29-oxa- 1,7,17-triazaoctacyclo[12.12.2.12,6.07,28.08,13.015,19.020,27.021,26]nonacosa-8,10,12,14,19,21,23,25,27-nonaen-16-one |
-7.7 |
|
55 |
CID 53230011 |
-7.1 |
|
56 |
Azalomycin |
-7.9 |
|
57 |
Schembl20768056 |
-8 |
|
58 |
(3Z,5Z)-3,5-Bis(3,4-dichlorobenzylidene)-1-(3-morpholinopropanoyl)-4-piperidone;hydrochloride |
-8.6 |
|
59 |
2-Amino-3-phosphino-propanoic acid |
-4.5 |
|
60 |
Sparsomycin |
-6.1 |
|
61 |
Triacsin c |
-4.8 |
Protein-ligand interaction analysis
The compounds showing higher binding energies (<-8 Kcal/mol) were selected and subjected to interaction analysis. The interaction of compounds on the binding sites of the target protein and nature of bonding were analyzed as shown in Table 4.
|
S. No |
Compound Name |
Protein-Ligand interaction |
|
|
|
|
No of H-bonds |
Binding Amino acid residues |
|
1 |
From Streptomyces tanashiensis |
2 |
H: 42-GLY, 110-THR |
|
2 |
Thaxtomine A |
3 |
H: 42-GLY, L: 39-LYS, 42-GLN |
|
3 |
Bafilomycin A1 from Streptomyces griseus |
3 |
H: 108-THR, L: 38-GLN, 39-LY |
|
4 |
Bafilomycin A1, Streptomyces griseus |
1 |
H: 148-GLU |
|
5 |
Borrelidin |
1 |
H: 42-GLY |
|
6 |
6,7,12,13-Tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one |
3 |
H: 39-GLN; 43-LYS, L: 165-GLU |
|
7 |
Kendomycin |
- |
- |
|
8 |
Antibiotic DC 107 |
2 |
H: 42-GLY, 110-THR |
|
9 |
Thaxtomin A |
3 |
H: 42-GLY, L: 39-LYS, 42-GLN |
|
10 |
Lactoquinomycin A |
2 |
H: 42-GLY, 110-THR |
|
11 |
Bafilomycin A1 |
1 |
H: 176-TYR |
|
12 |
Leinamycin |
2 |
H: 42-GLY, 110-THR |
|
13 |
Schembl20768056 |
5 |
H: 42-GLY, 91-TYR, 148-GLU, L: 38-GLN |
|
14 |
(3Z,5Z)-3,5-Bis(3,4-dichlorobenzylidene) -1-(3-morpholinopropanoyl)-4-piperidone; hydrochloride LYS |
3 |
H: 108_THR, L: 41-GLY, 103- |
Figures 4 to 17 show the results of the finalized compounds below:
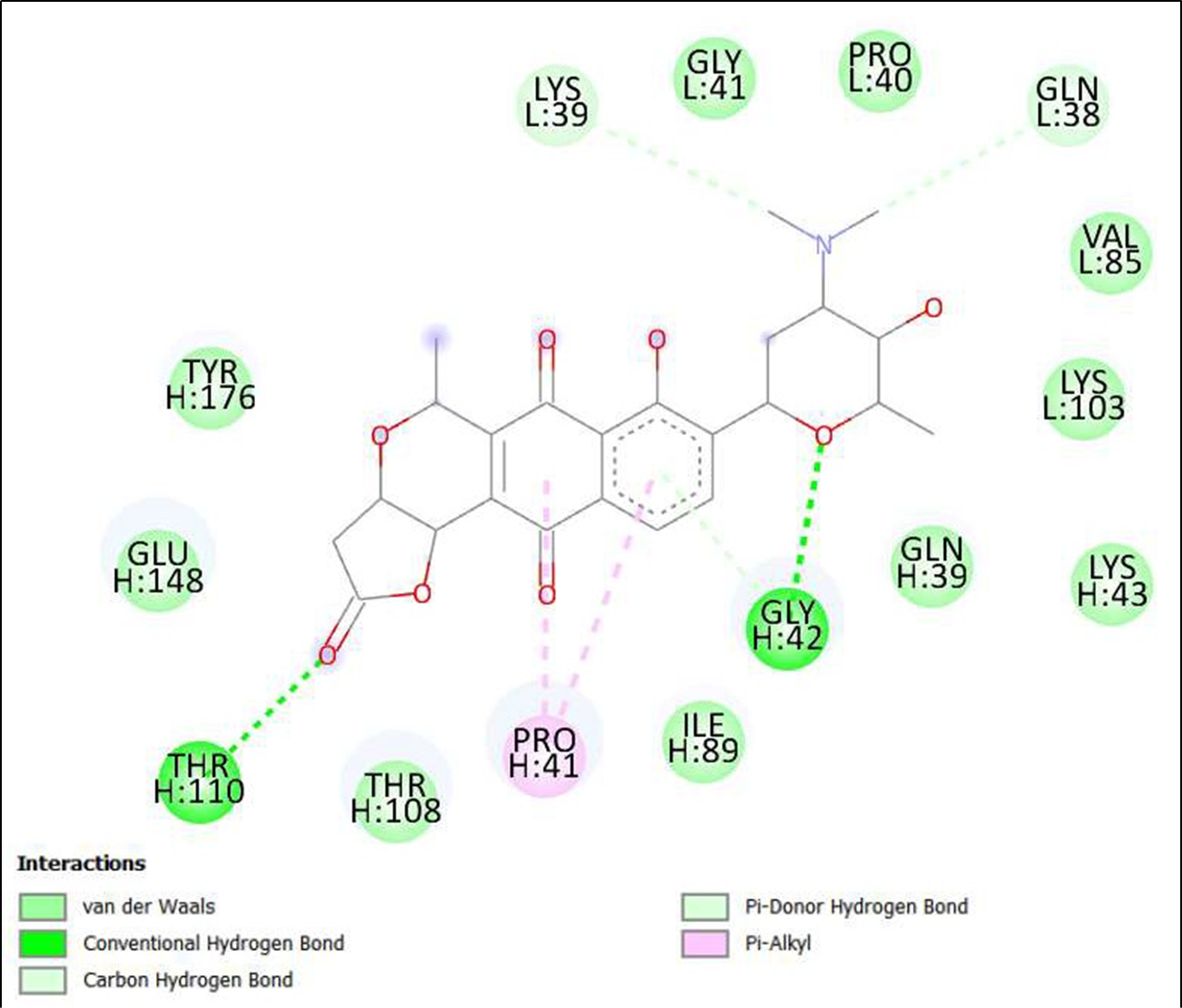
Figure 4. Interaction of compounds From Streptomyces tanashiensis on binding sites of SARS-CoV-2 receptor binding domain (RBD).
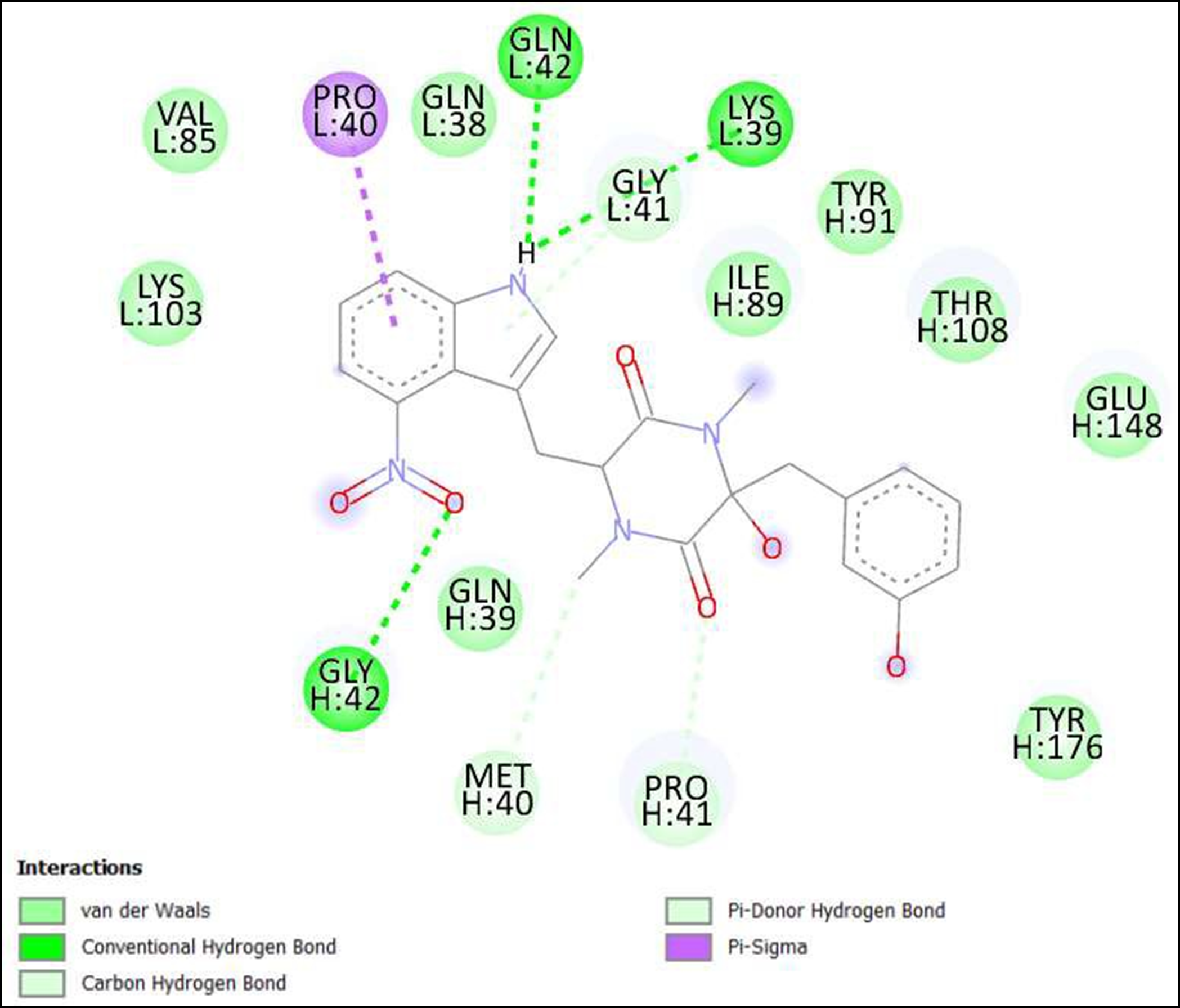
Figure 5. Interaction of Thaxtomine A on binding sites of SARS-CoV-2 receptor binding domain (RBD).
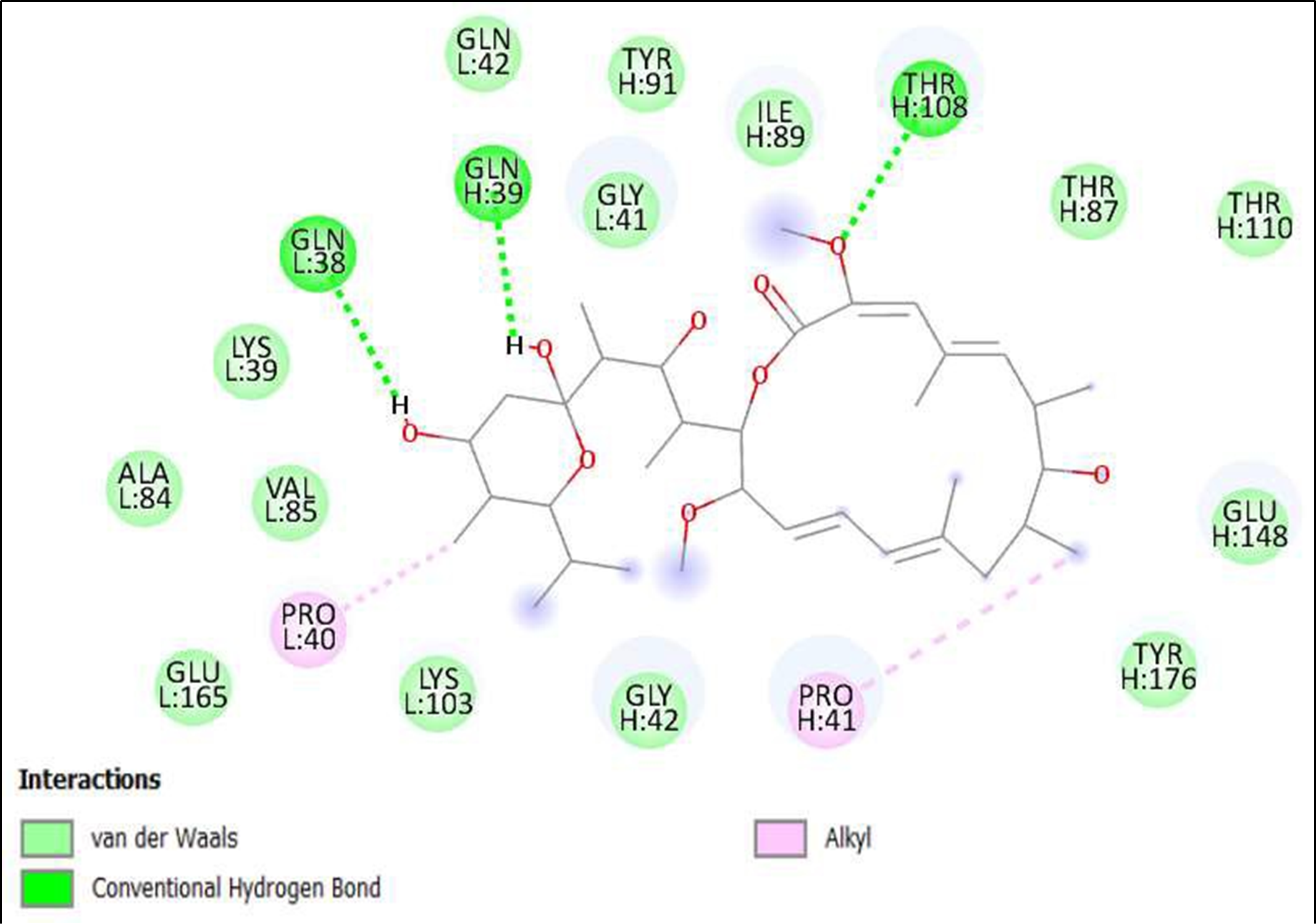
Figure 6. Interaction of Bafilomycin A1 from Streptomyces griseus on binding sites of SARS-CoV-2 receptor binding domain (RBD).
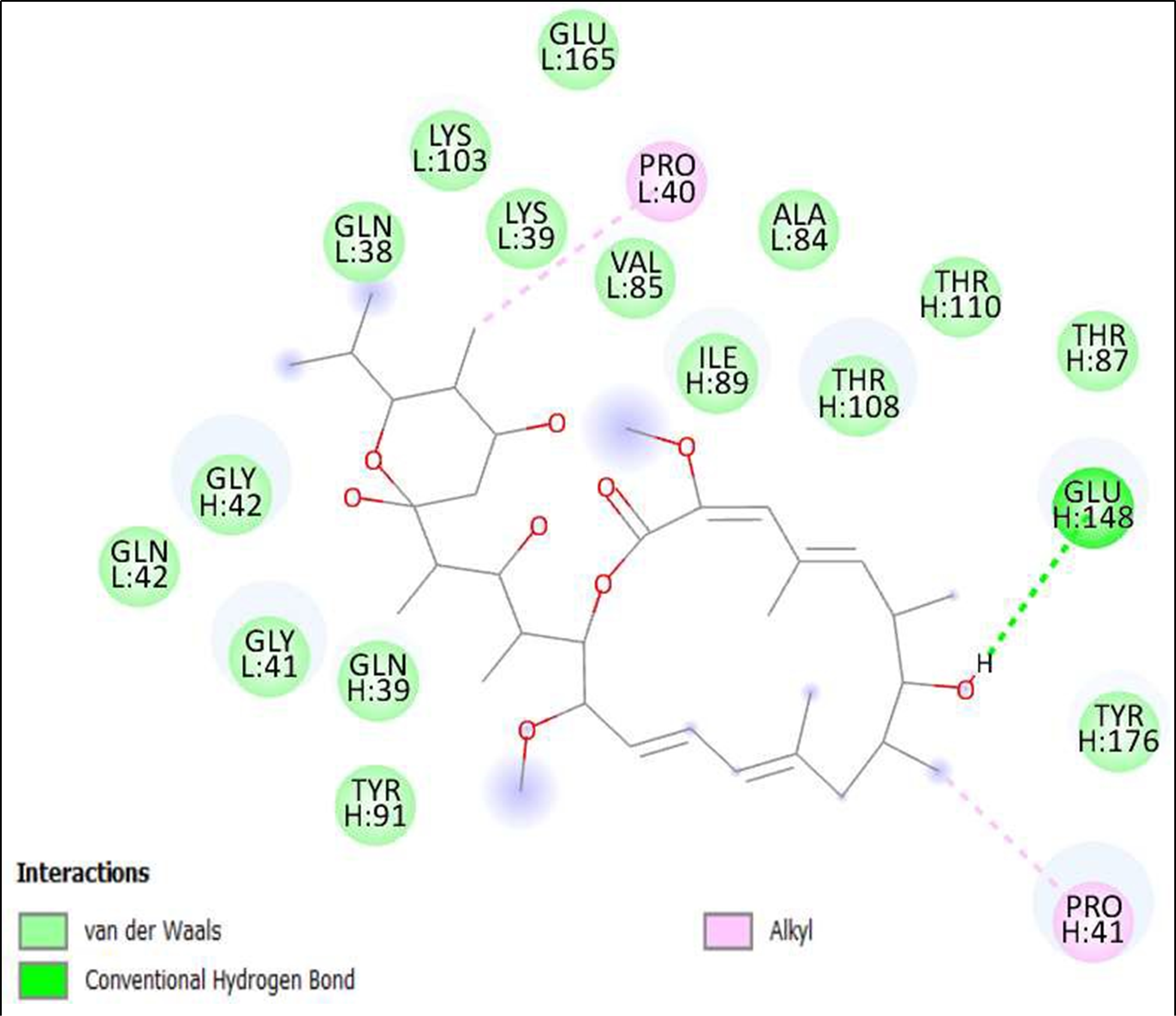
Figure 7. Interaction of Bafilomycin A1, Streptomyces griseus on binding sites of SARS-CoV-2 receptor binding domain (RBD).
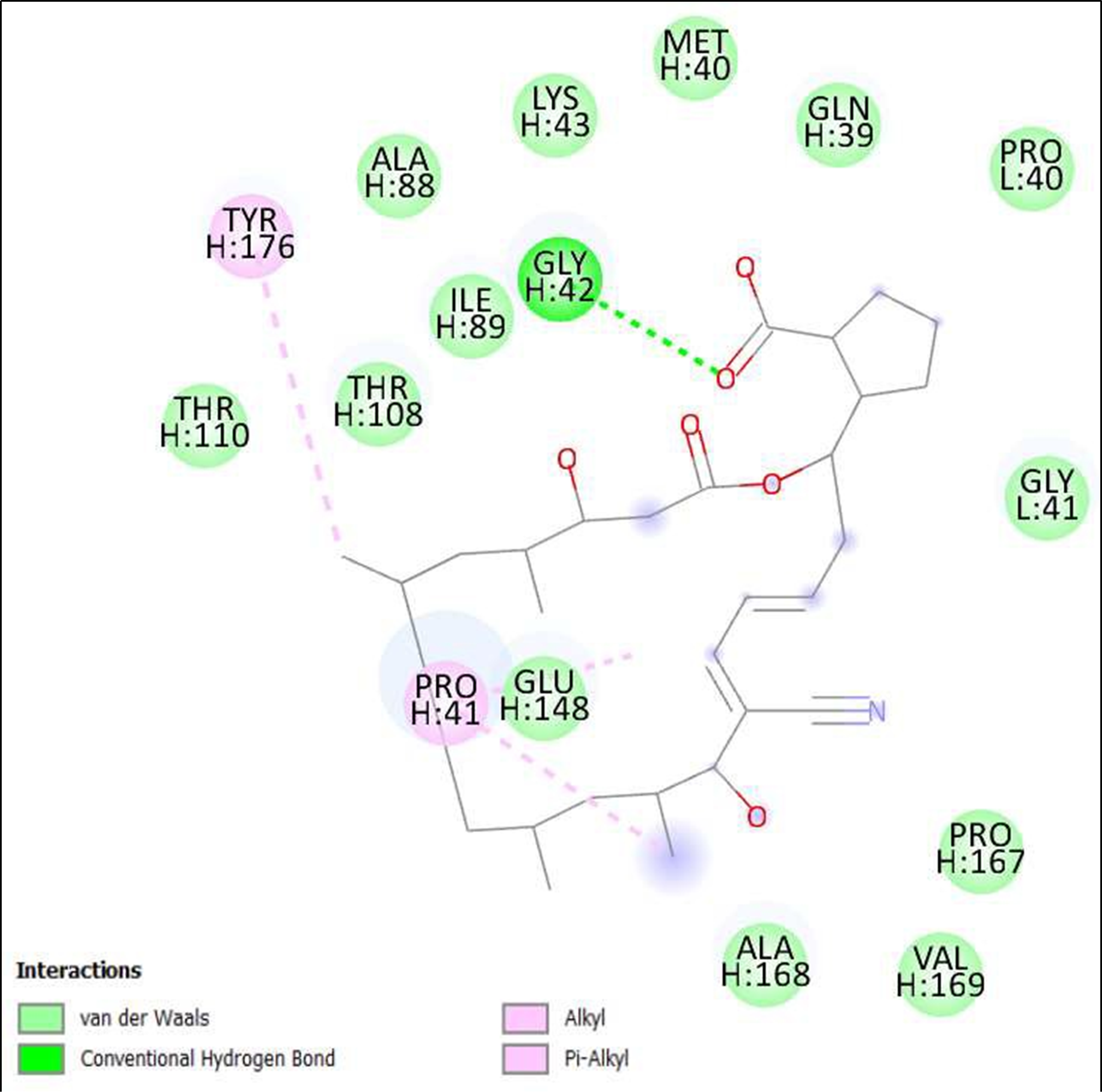
Figure 8. Interaction of Borrelidin on binding sites of SARS-CoV-2 receptor binding domain (RBD).
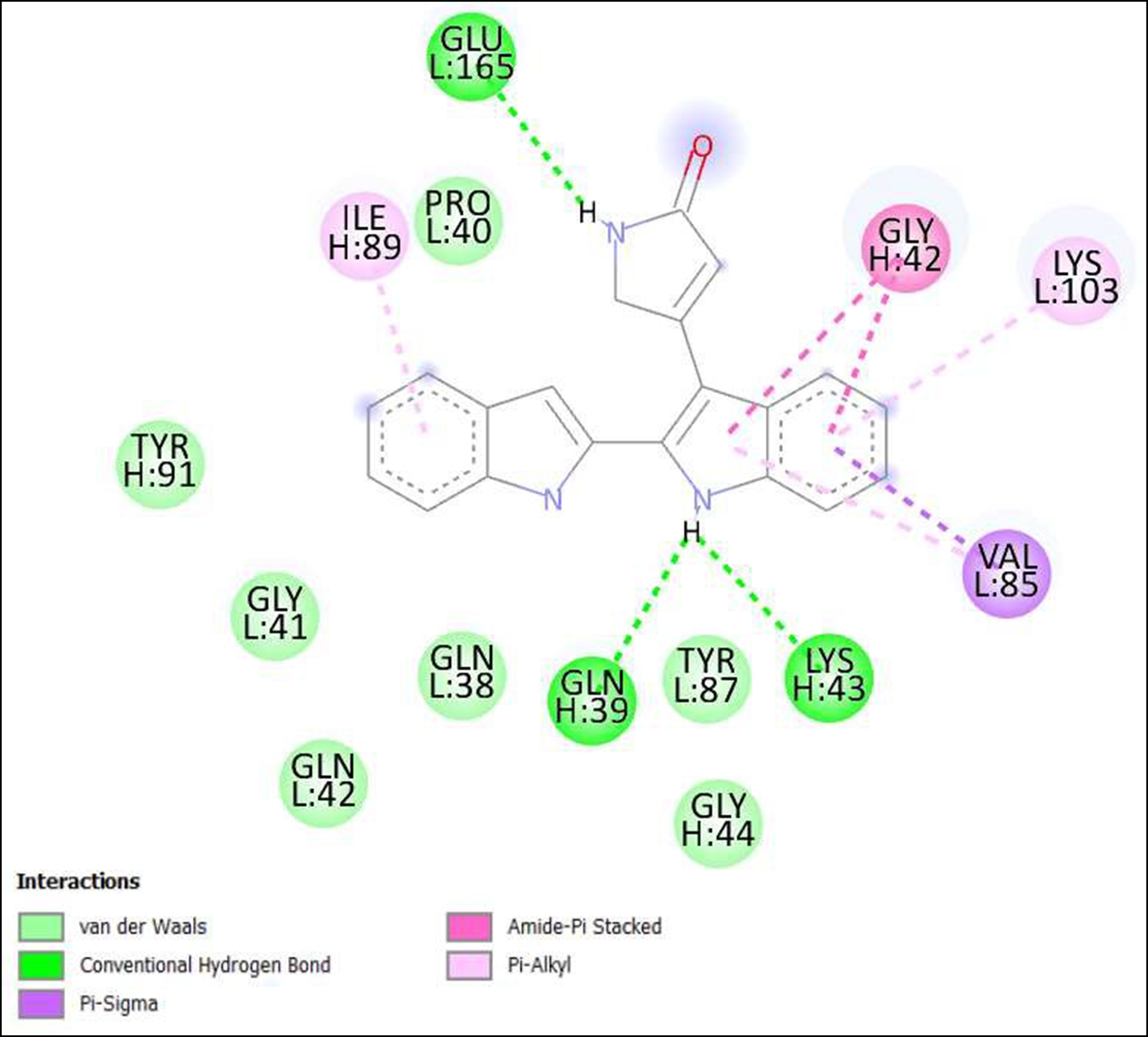
Figure 9. Interaction of 6,7,12,13-Tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one on binding sites of SARS-CoV-2 receptor binding domain (RBD).
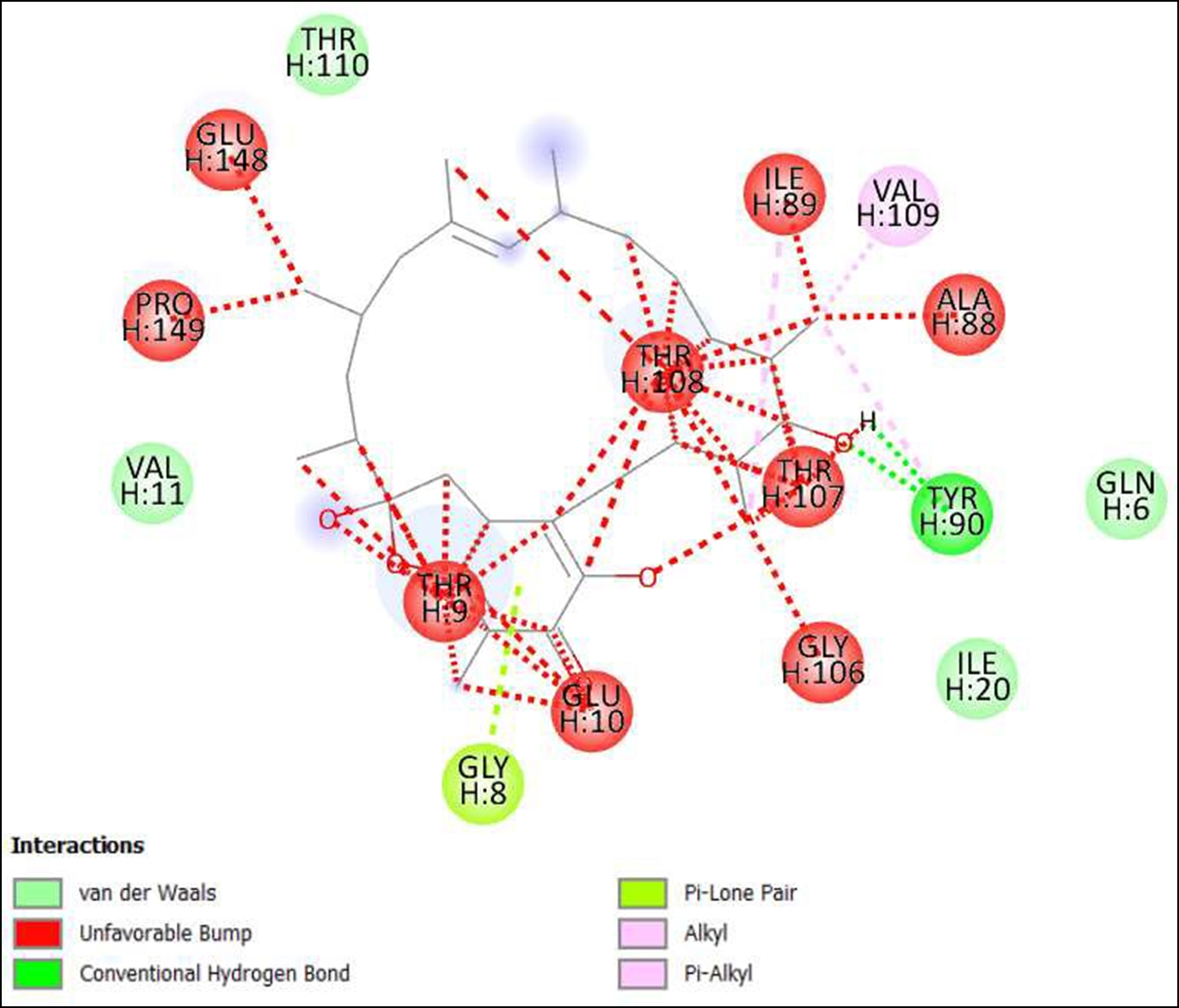
Figure 10. Interaction of Kendomycin on binding sites of SARS-CoV-2 receptor binding domain (RBD).
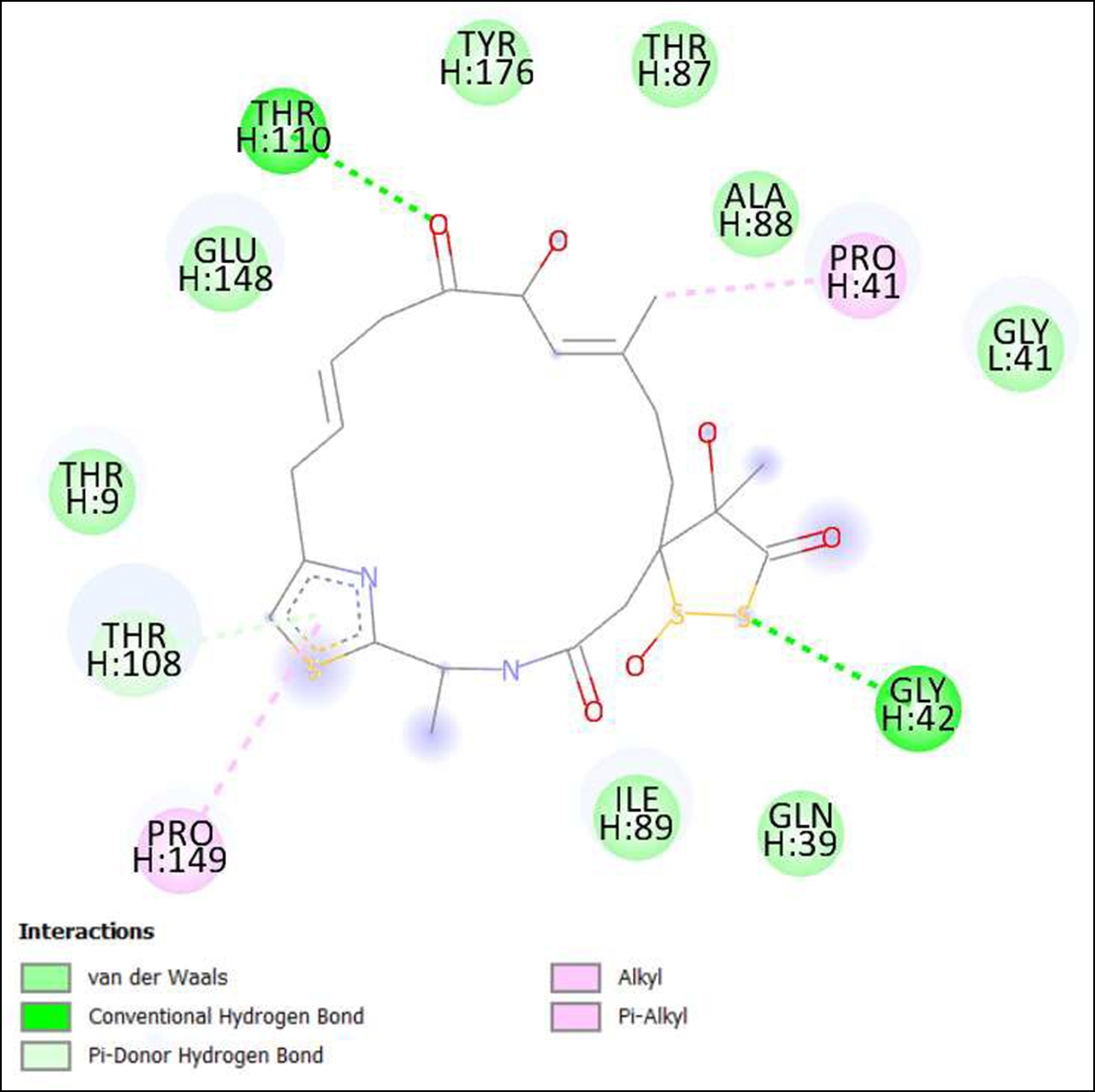
Figure 11. Interaction of Antibiotic DC 107 on binding sites of SARS-CoV-2 receptor binding domain (RBD).
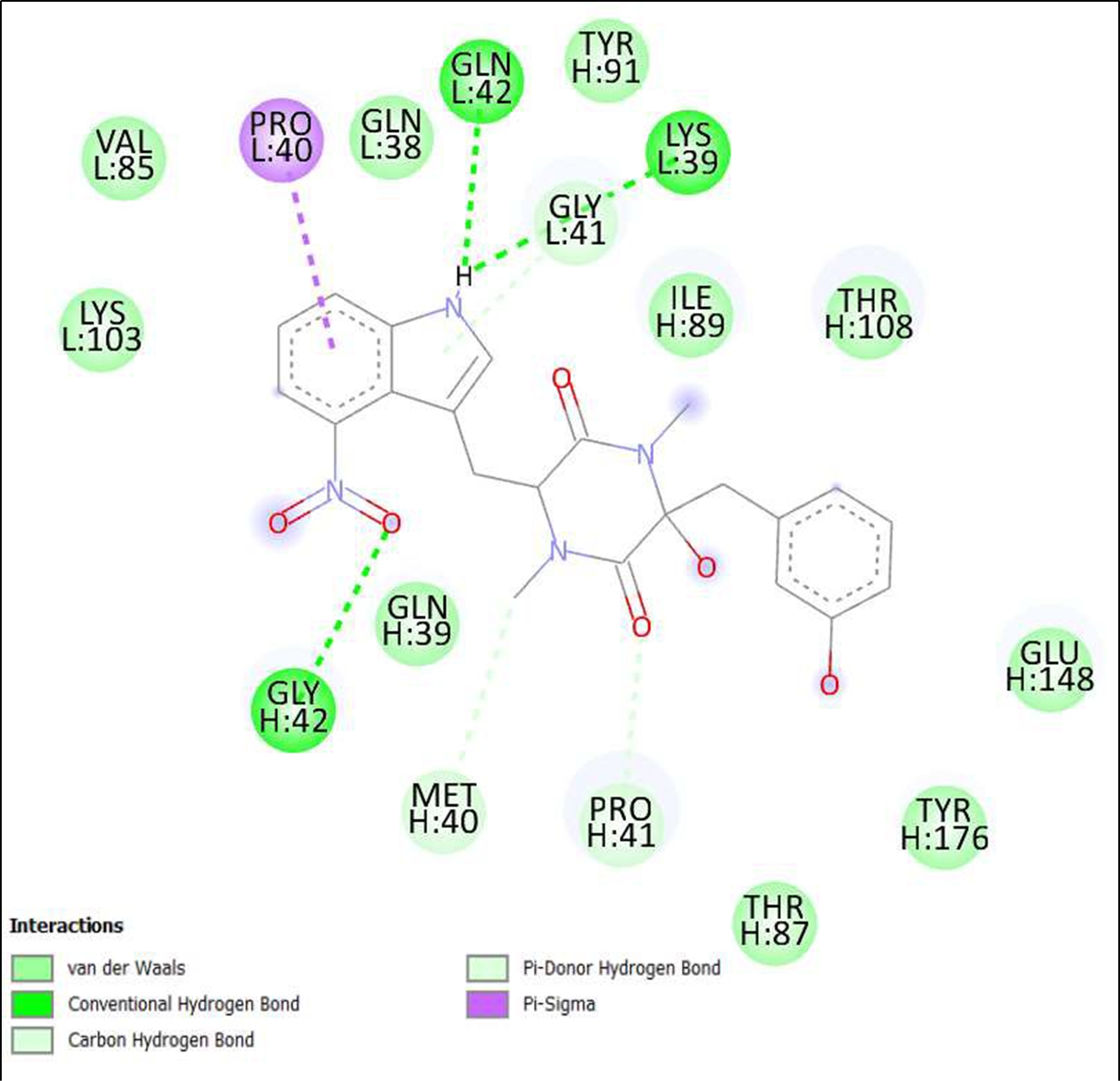
Figure 12. Interaction of Thaxtomin A on binding sites of SARS-CoV-2 receptor binding domain (RBD).
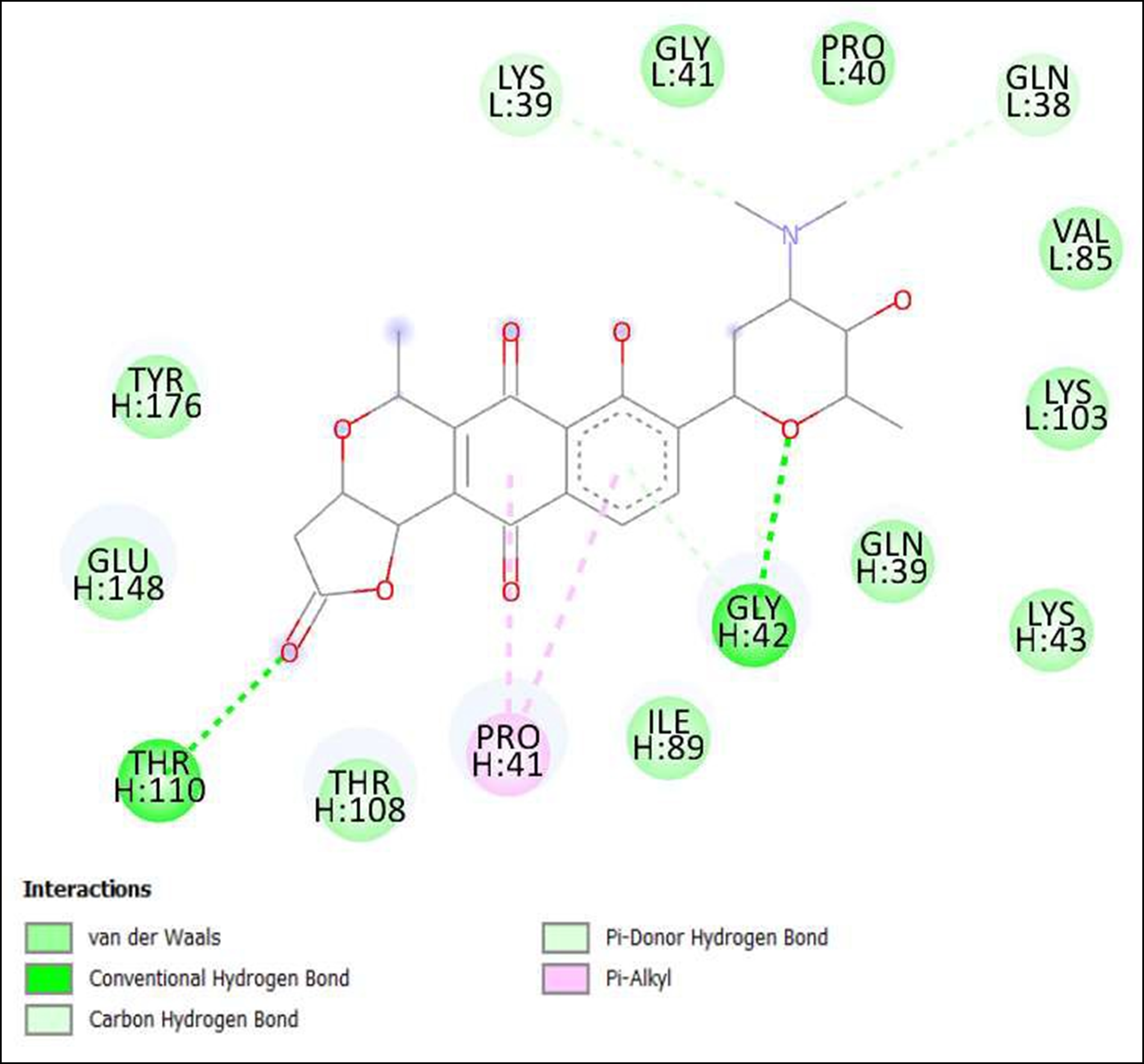
Figure 13. Interaction of Lactoquinomycin A on binding sites of SARS-CoV-2 receptor binding domain (RBD).
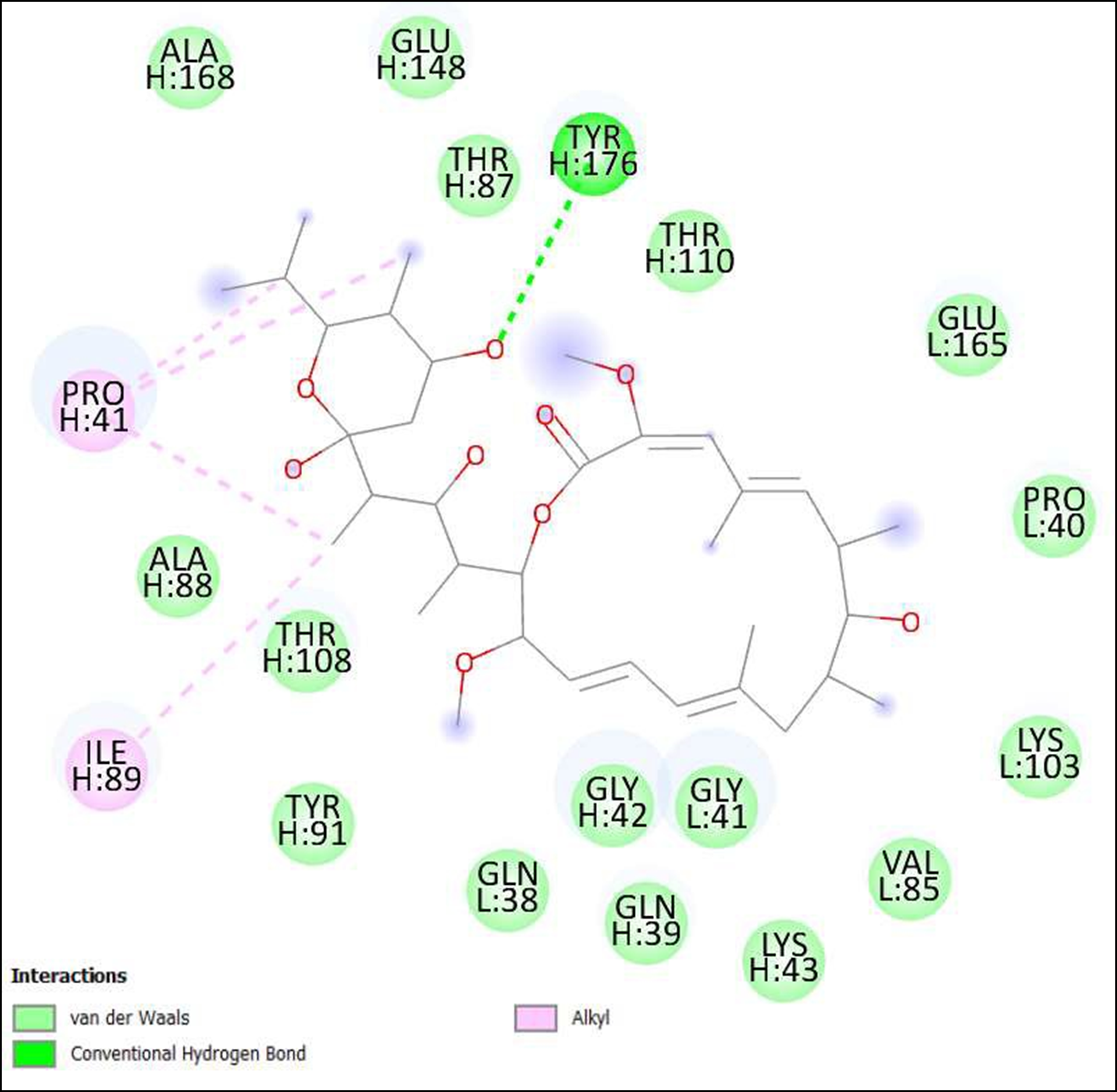
Figure 14. Interaction of Bafilomycin A1 on binding sites of SARS-CoV-2 receptor binding domain (RBD).
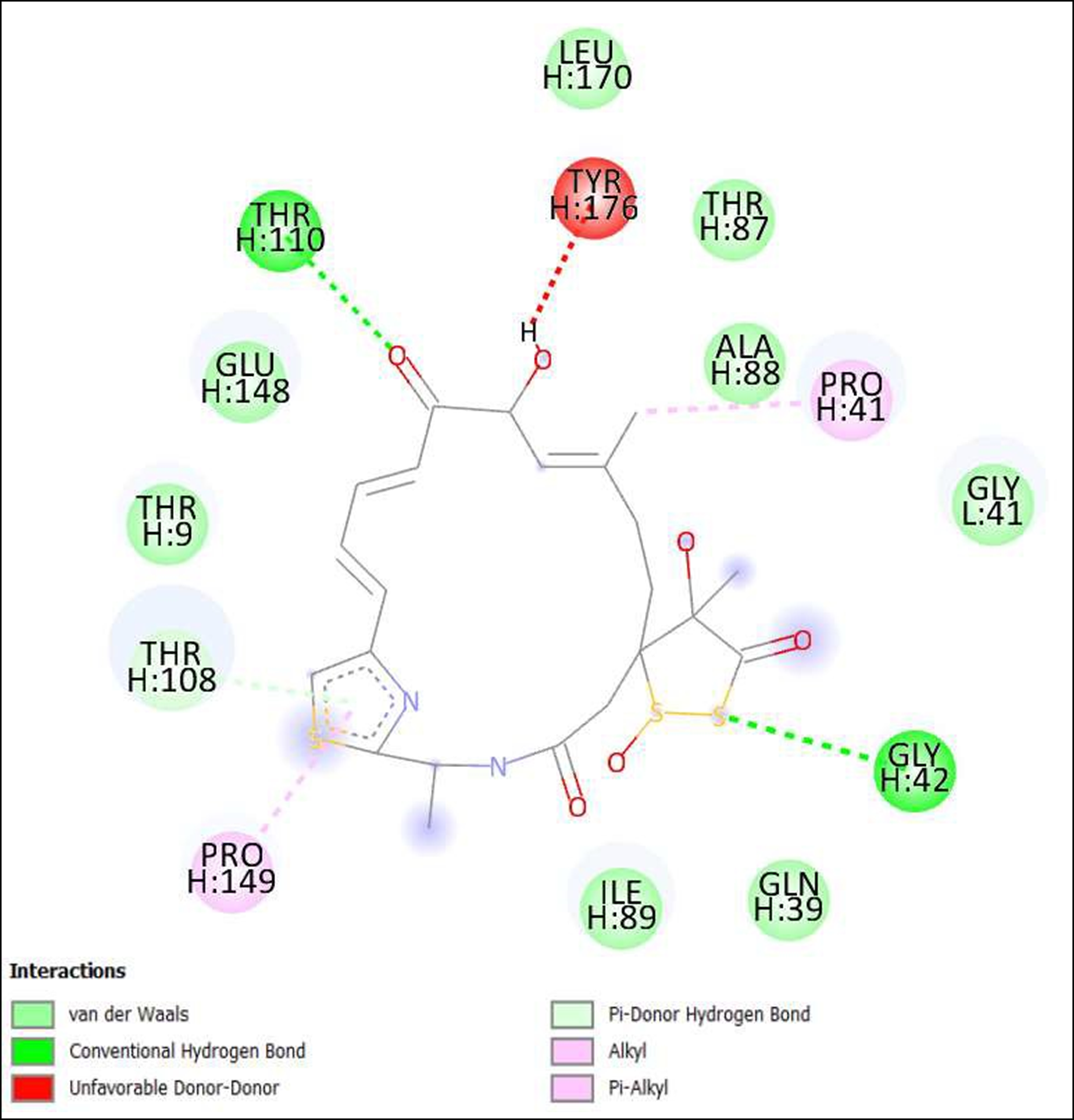
Figure 15. Interaction of Leinamycin on binding sites of SARS-CoV-2 receptor binding domain (RBD).
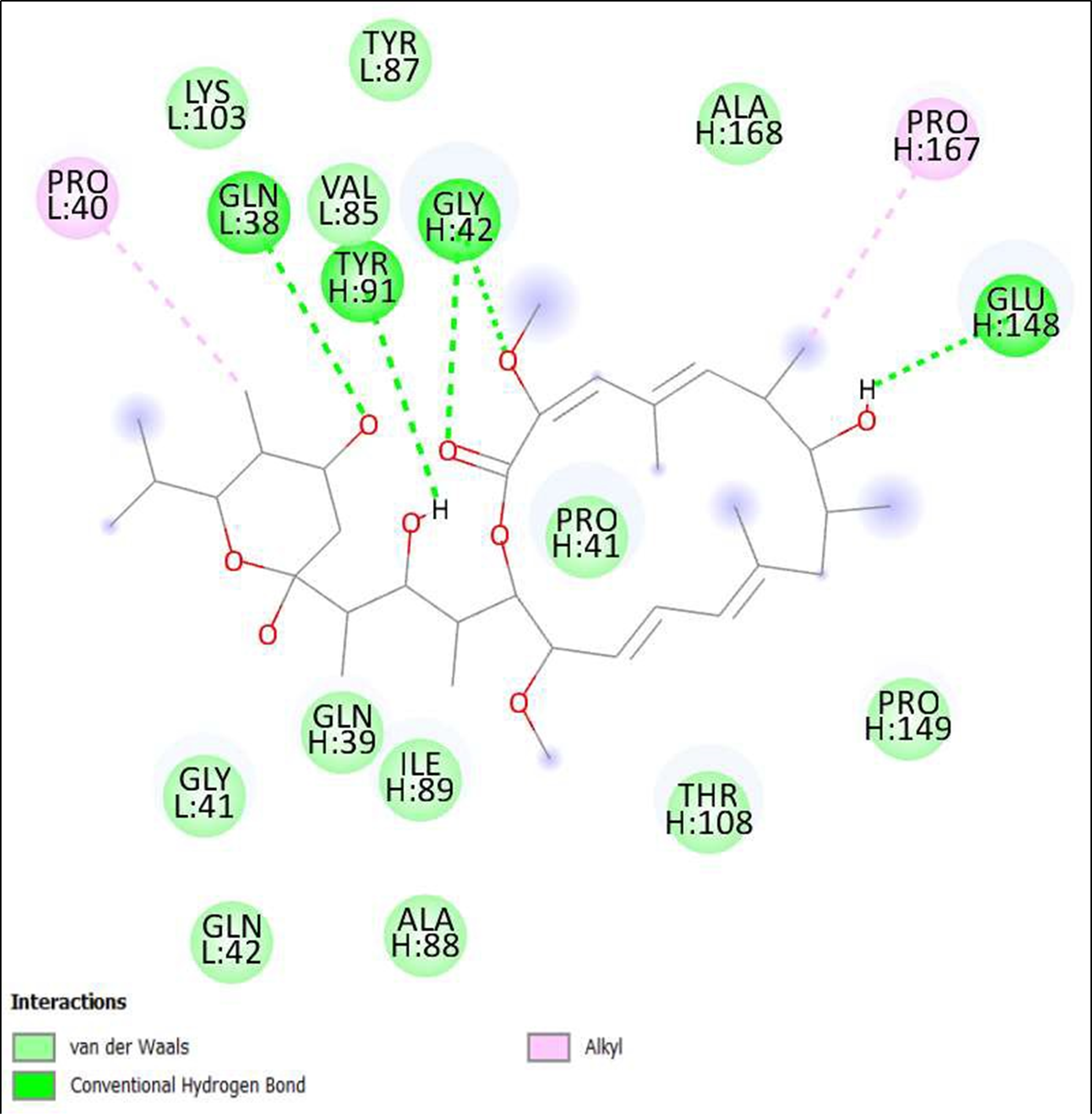
Figure 16. Interaction of Schembl20768056 on binding sites of SARS-CoV-2 receptor binding domain (RBD).
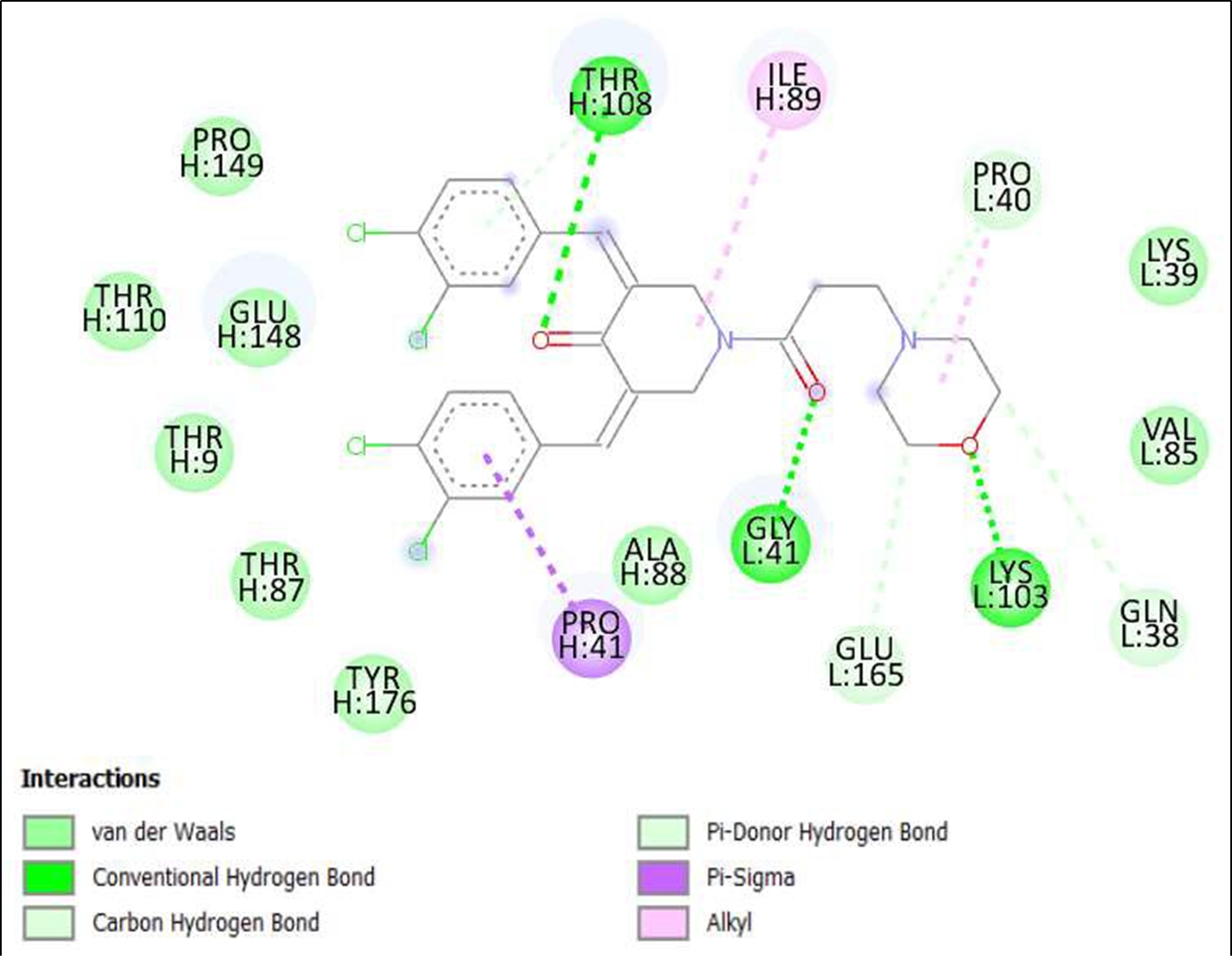
Figure 17. Interaction of (3Z,5Z)-3,5-Bis(3,4-dichlorobenzylidene)- 1-(3-morpholinopropanoyl)-4- piperidone; hydrochloride on binding sites of SARS-CoV-2 receptor binding domain (RBD).
Discussion
According to research papers published and studies conducted worldwide, few of the compounds that were used in drug development of COVID are enlisted below with their test results -
Escherichia coli - This bacterium is typically utilized to manufacture recombinant protein and is used in production of antibodies and vaccines such as Pfizer-BioNTech and Moderna. Vaccine candidates frequently contain the COVID-19 spike protein RBD. Due to the small molecular mass along with their structural rearrangement of full-length SP and detachment, the RBD's immunogenicity is rather limited [9].
Saccharomyces cerevisiae - In immunocompromised and critically ill patients, the saccharomyces organisms are reported as agents of invasive infection. The antitoxin of MazE i.e. MazF along with the toxin itself from Escherichia coli were combined with a protease cleavage site in the linker peptide. The viral protease cleaves the MazEF fusion, liberating MazF toxin from its antitoxin and inhibiting growth. A fluorescent marker protein on the modified yeast strain lets track its development with or without inhibitors [10].
CHO cells - Biopharmaceuticals are often made using CHO cells. A recombinant vaccine was made using a prefusion-stabilized SARS-CoV-2 spike trimer and aluminum hydroxide. CHO cells were used to isolate and stabilize spike protein. In mice and non- human primates, candidate vaccinations strongly induced CD4+ T cell and neutralizing antibody responses [11].
HEK293 cells - Genetically engineered human embryonic kidney cells produce protein efficiently. They manufacture COVID-19 vaccines and antibodies for possible therapies. SARS-CoV infection induced CPE, and quantitative real-time polymerase chain reaction measured viral replication. Indirect immunofluorescence confirmed. 10 SARS-CoV- susceptible cell lines were found. SARS-CoV replication and CPE occurred in HEK-293 cells [12].
African green monkey kidney (vero) cells - Sequencing revealed that influenza B viruses formed from Vero cells differ from egg-grown viruses at amino acids 196–198 but share a hemagglutinin with MDCK-grown viruses. Fluorescence-activated cell sorting shows that human influenza A and B viruses can infect Vero cells with alpha2,3 galactose-linked sialic acid. Electron microscopy and immunofluorescence showed that infected Vero cells become polarized epithelia. Vero cells can generate pandemic-vaccinating influenza A and B viruses [13].
Bacillus subtilis - Inactivated B. subtilis spores increase neutralizing antibody levels for weeks following booster treatment. The inactivated oral vaccination booster candidate's immediate effects are predicted to elicit gut mucosal immune responses simply by the spore surface, followed by systemic effects. When neutralizing antibodies decline and stabilize after 6 months, Bacillus subtilis is the key component for booster doses [14].
Lactococcus lactis - L. lactis NZ3900 expressed the HCR spike protein gene from pNZ8149. ELISA evaluated anti-SARS-CoV2 spike antibody (IgG and IgA) levels before and after therapy. Plasma and lymph and intestinal CD4+ and CD8+ lymphocytes were examined. L. lactis bearing the HCR gene raised IgG/IgA-SARS-CoV-2 levels in experimental mice after nasal and oral treatment [15].
Pichia pastoris - Heterologous protein expression in vaccine development is best in Pichia pastoris (P. pastoris). It reduces production costs by combining the speed and convenience of highly efficient prokaryotic platforms with certain mammalian system characteristics. Low- and middle-income countries produced vaccines cheaply [16].
Tobacco and Tomato Plants - Vaccines, pharmaceuticals, immunomodulatory proteins, and monoclonal antibodies are bioreactive or factory products produced from plants with the help of molecular farming/transient expression systems. These biological products are stable, safe, efficacious, inexpensive, and readily available. Plant molecular farming might speed up industrial biologics manufacturing in emergencies like the COVID-19 pandemic [17].
Aspergillus niger and Aspergillus oryzae - Coronavirus-associated pulmonary aspergillosis (CAPA) may cause morbidity and death in COVID-19 patients, while many elements of the illness are yet unknown. Endogenous immunomodulation may restore immunological homeostasis in COVID-19 patients and minimize aspergillosis risk. COVID-19's opposite side is CAPA [18].
Chlorella vulgaris and Chlamydomonas reinhardtii - To test an oral algae-based vaccination, mice were given a Chlamydomonas-made SARS-CoV-2 RBD. In freeze-dried algal biomass, the test immunogens were quite stable in nature and systemic and mucosal humoral responses against the spike protein by mouth at similar levels as injected antigen + alum adjuvant were produced. IgG subclass analysis showed a Th2-bias response lasting over 4 months following the previous vaccination. Induced antibodies reacted similarly to Delta and Omicron versions [19].
Haloferax volcanii - Halophilic enzymes may be useful in bio-catalysis, biological remediation, bio-plastics, nano-biotechnology, and biofuels. A wide genetic toolbox with possibilities for controlled protein overexpression has allowed H. volcanii to purify biotechnologically significant enzymes from diverse halophiles. New biotechnology applications could be made possible by the use of expressed active proteins in immobilized cells in a porous biocompatible matrix for H. volcanii [20].
Call free systems - Cell-free protein synthesis (CFPS) systems, which utilize removed transcriptional and translational machinery from cells, may rapidly create proteins without the restrictions of living cells and be used for on-demand biomanufacturing. CFPS systems have been used to produce subunit, conjugate, virus-like particle (VLP), and membrane-augmented vaccinations and design vaccines [21].
Baculoviruses - Baculovirus expression vectors express glycoproteins, enzymes, vaccines, and recombinant viruses. Non-Structural Proteins (NSPs), another recombinant SARS-CoV-2 protein produced by BEV, are utilized to find new COVID-19 treatments or repurpose current ones. BEV produces therapeutic proteins, recombinant antibodies, MMLVRT, and ACE2 for SARS-CoV-2 diagnostic and therapy. Modified recombinant proteins increased production or stability [22].
Pseudomonas fluorescens - Gram-negative bacteria's ubiquitous transcription regulator Ferric uptake regulator (Fur) controls several biological activities. The pathogenic Pseudomonas fluorescens strain TSS, which was acquired from locally farmed Japanese flounder, was used to clone the fur gene. Escherichia coli fur mutants can partially benefit from TSS Fur [23].
Streptomyces lividans - Streptomyces coelicolor and Mycobacterium TB' genomes imply a shared Actinomycete ancestry. Streptomyces vaccination can be used as a live vector and Mycobacterial infection protecter. The theoretical proteomes of S. coelicolor A3(2) and M. tuberculosis H37Rv and Mycobacterium bovis AF2122/97 exhibited substantial gene sequence similarity and shared membrane proteins. This bacterium produces a COVID-19- diagnostic protein [24].
Lactic acid bacteria - Gram-positive, nonpathogenic recombinant lactic acid bacteria (LAB) strains induce particular systemic and mucosal immune responses against chosen antigens. Modern vaccinology prioritizes vaccine design, mucosally administered vaccines, in vivo antigen production monitoring, optimal vaccine dosage, strain selection, and characterization [25].
Mycobacterium smegmatis - SARS-spike CoV-2 and nucleocapsid are carried by M. smegmatis. S or N antigens were fused with Mycobacterium TB cell wall trafficking component PE PGRS33 protein to mimic viral particle surface localization (M.tb). After receiving the COVID-19 vaccine candidate based on M. smegmatis, mice developed CD4+ and CD8+ T cells with central memory as well as S or N antigen-specific T cell immune responses. In T cell-based vaccines for SARS-CoV-2, M. smegmatis may be utilized [26].
It was observed that only one compound Streptomyces lividans was effectively utilized for drug development even though Streptomyces has been a proven antibiotic of natural origin.
Lee et al. used genome sequences for mining novel secondary metabolite biosynthetic gene clusters [27]. Senges, et al. [28] used the technique metabolomics. In Ouchene et al. study [29], integrated metabolomics, molecular networking and genome mining analysis was used. In Iglesias et al. performed identification, characterization and in-silico analysis in their studies [30]. Edison et al. [31] used in-silico analysis in structural elucidation. Some researchers used comparative genomics in their studies [32-37]. Kumar et al. [38] used in vitro and in-silico analysis using machine learning in their studies. It was observed that genome mining, metabolomics, structural modeling, comparative genomics, and machine learning were the most used in-silico techniques with Streptomyces.
In this research, in-silico technique is used and the following steps were used to perform the complete process. The first step being the Selection and Preparation of Ligands followed by Screening of these Ligands based on the Druglikeness. The last few steps to complete the entire process are Preparation of Target proteins with identifying the binding sites and the final step being the Molecular docking and Protein Ligand interaction analysis to obtain the end results of the study. All these steps play a crucial role for the entire process. The first step allows one to identify and select the potential ligands which would be useful for the study from multiple databases. Screening on the basis of druglikeness helps in identifying whether the potential ligands selected would be a good candidate for the development of drugs on the basis of their physicochemical properties. Preparation of target proteins and identifying the binding sites aid in the successfulness of molecular docking simulations. These also improve the accuracy and lower the chances of getting false positive results. The final step is used to predict the binding affinity and the mode of interaction between ligands with their target proteins. Before a compound is synthesized and evaluated in vitro or in vivo, this can provide invaluable information about its potential efficacy and safety.
Streptomyces is a type genus of the Streptomycetaceae family and presently encompasses close to 576 species, with the number growing annually. There was a significant scope for research; and hence this research work lays a groundwork focused on discovering the other potential compounds that can further be used for the drug development for the novel virus.
Conclusion
From a total of 190, compounds were screened based on drug likeness analysis. 61 compounds showed drug likeness properties and hence were further subjected to docking studies. The compounds showing higher binding energies (<-8 Kcal/mol) were utilized for interaction analysis. All the 14 compounds except Kendomycin showed interaction on binding sites of target proteins. Therefore, the compounds identified from the study i.e. From Streptomyces tanashiensis, Thaxtomine A, Bafilomycin A1 from Streptomyces griseus; Bafilomycin A1, Streptomyces griseus; Borrelidin, 6,7,12,13-Tetrahydro-5H-indolo[2,3- a]pyrrolo[3,4-c]carbazol-5-one, Antibiotic DC 107, Thaxtomin A, Lactoquinomycin A, Bafilomycin A1, Leinamycin, Schembl20768056, (3Z,5Z)-3,5-Bis(3,4- dichlorobenzylidene)-1-(3-morpholinopropanoyl)-4-piperidone, hydrochloride can be used for drug development for treatment of COVID-19 infections. Further studies will confirm the proper usage and implementation of new compounds for procuring the COVID-19 antivirals.
Acknowledgement
The authors are thankful to the Darwin Conference for giving them an opportunity for presenting the following research and grateful to them as well as Scientific Archives for publishing this work into their journals.
Funding Statement
No funding received for this research work.
Conflict of Interest
There are no conflicts of interest pertaining to this paper by the authors.
References
2. Euzéby JP, Parte AC. "Streptomyces". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved June 9, 2021.
3. van der Meij A, Willemse J, Schneijderberg MA, Geurts R, Raaijmakers JM, van Wezel GP. Inter- and intracellular colonization of Arabidopsis roots by endophytic actinobacteria and the impact of plant hormones on their antimicrobial activity. Antonie Van Leeuwenhoek. 2018 May;111(5):679-690.
4. Kämpfer P. The Family Streptomycetaceae, Part I: Taxonomy. In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds.). The Prokaryotes. 2006; pp. 538– 604.
5. Madigan M, Martinko J, eds. Brock Biology of Microorganisms (11th ed.). Prentice Hall. 2005.
6. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics (2nd ed.). Norwich, England: John Innes Foundation. 2000.
7. Bibb MJ. Understanding and manipulating antibiotic production in actinomycetes". Biochemical Society Transactions. 2013 Dec; 41(6):1355-64.
8. https://eol.org/pages/83069/articles?locale_code=en#cite_noteBrock-5
9. Bellone ML, Puglisi A, Dal Piaz F, Hochkoeppler A. Production in Escherichia coli of recombinant COVID-19 spike protein fragments fused to CRM197. Biochem Biophys Res Commun. 2021 Jun 18;558:79-85.
10. Alalam H, Sigurdardóttir S, Bourgard C, Tiukova I, King RD, Grøtli M, et al. A Genetic Trap in Yeast for Inhibitors of SARS-CoV-2 Main Protease. mSystems. 2021 Dec 21;6(6):e0108721.
11. Liu H, Zhou C, An J, Song Y, Yu P, Li J, et al. Development of recombinant COVID-19 vaccine based on CHO-produced, prefusion spike trimer and alum/CpG adjuvants. Vaccine. 2021 Nov 26;39(48):7001-7011.
12. Kaye M. SARS-associated coronavirus replication in cell lines. Emerg Infect Dis. 2006 Jan;12(1):128-33.
13. Govorkova EA, Murti G, Meignier B, de Taisne C, Webster RG. African green monkey kidney (Vero) cells provide an alternative host cell system for influenza A and B viruses. J Virol. 1996 Aug;70(8):5519- 24.
14. Sung JC, Lai NC, Wu KC, Choi MC, Ma CH, Lin J, et al. Safety and Immunogenicity of Inactivated Bacillus subtilis Spores as a Heterologous Antibody Booster for COVID-19 Vaccines. Vaccines (Basel). 2022 Jun 24;10(7):1014.
15. Yurina V, Rahayu Adianingsih O, Widodo N. Oral and intranasal immunization with food-grade recombinant Lactococcus lactis expressing high conserved region of SARS- CoV-2 spike protein triggers mice's immunity responses. Vaccine X. 2023 Apr;13:100265.
16. de Sá Magalhães S, Keshavarz-Moore E. Pichia pastoris (Komagataella phaffii) as a Cost-Effective Tool for Vaccine Production for Low- and Middle-Income Countries (LMICs). Bioengineering (Basel). 2021 Aug 31;8(9):119.
17. Dhama K, Natesan S, Iqbal Yatoo M, Patel SK, Tiwari R, Saxena SK, Harapan H. Plant- based vaccines and antibodies to combat COVID-19: current status and prospects. Hum Vaccin Immunother. 2020 Dec 1;16(12):2913-2920.
18. Costantini C, van de Veerdonk FL, Romani L. Covid-19-Associated Pulmonary Aspergillosis: The Other Side of the Coin. Vaccines (Basel). 2020 Dec 1;8(4):713.
19. Govea-Alonso DO, Malla A, Bolaños-Martínez OC, Vimolmangkang S, Rosales- Mendoza S. An Algae-Made RBD from SARS-CoV-2 Is Immunogenic in Mice. Pharmaceuticals (Basel). 2022 Oct 21;15(10):1298.
20. Haque RU, Paradisi F, Allers T. Haloferax volcanii for biotechnology applications: challenges, current state and perspectives. Appl Microbiol Biotechnol. 2020 Feb;104(4):1371-1382.
21. Hu VT, Kamat NP. Cell-free protein synthesis systems for vaccine design and production. Curr Opin Biotechnol. 2023 Feb;79:102888.
22. Azali MA, Mohamed S, Harun A, Hussain FA, Shamsuddin S, Johan MF. Application of Baculovirus Expression Vector system (BEV) for COVID-19 diagnostics and therapeutics: a review. J Genet Eng Biotechnol. 2022 Jul 6;20(1):98.
23. Wang HR, Hu YH, Zhang WW, Sun L. Construction of an attenuated Pseudomonas fluorescens strain and evaluation of its potential as a cross-protective vaccine. Vaccine. 2009 Jun 19;27(30):4047-55.
24. Arzuaga NO, Vila Granda A, Gómez JC, San Miguel ME, Bourzac JF, Hernández YL, Elías López AL, Pólux CR, Mesa LG, HernándezPando R, Domínguez AA. The use of Streptomyces for immunization against mycobacterial infections. Hum Vaccin. 2011 Sep;7(9):934-40.
25. Szatraj K, Szczepankowska AK, Chmielewska-Jeznach M. Lactic acid bacteria - promising vaccine vectors: possibilities, limitations, doubts. J Appl Microbiol. 2017 Aug;123(2):325-339.
26. Wen Z, Fang C, Liu X, Liu Y, Li M, Yuan Y, Han Z, Wang C, Zhang T, Sun C. A recombinant Mycobacterium smegmatis-based surface display system for developing the T cell-based COVID-19 vaccine. Hum Vaccin Immunother. 2023 Dec 31;19(1):2171233.
27. Lee N, Kim W, Hwang S, Lee Y, Cho S, Palsson B, et al. Thirty complete Streptomyces genome sequences for mining novel secondary metabolite biosynthetic gene clusters. Scientific Data. 2020;7:55.
28. Senges CHR, Al-Dilaimi A, Marchbank DH, Wibberg D, Winkler A, Haltli B, et al. The secreted metabolome of Streptomyces chartreusisand implications for bacterial chemistry. Proceedings of the National Academy of Sciences. 2018;115(10):2490-2495.
29. Ouchene R, Stien D, Segret J, Kecha M, Rodrigues AMS, Veckerlé C, et al. Integrated Metabolomic, Molecular Networking, and Genome Mining Analyses Uncover Novel Angucyclines From Streptomyces sp. RO-S4 Strain Isolated From Bejaia Bay, Algeria. Front Microbiol. 2022 Jun 23;13:906161.
30. Iglesias C, Tijman A, Lopez G, Pianzzola MJ, Panizza P, Giordani SR. Identification, Characterization, and In Silico Analysis of New Imine Reductases From Native Streptomyces Genomes. Frontiers in Catalysis. 2021;1:785963.
31. Edison LK, Reji SR, Pradeep NS. In silico perceptions in structural elucidation of Exo-ß-1, 4-glucanase and Endo-ß-1, 3-glucanase from Streptomyces spp. Journal of Applied Biology and Biotechnology. 2020; 8(6):67-81.
32. Alam K, Islam MM, Islam S, Hao J, Abbasi MN, Hayat M, et al. Comparative genomics with evolutionary lineage in Streptomyces bacteria reveals high biosynthetic potentials. World J Microbiol Biotechnol. 2022 Dec 30;39(2):64.
33. Malik A, Kim YR, Jang IH, Hwang S, Oh DC, Kim SB. Genome-based analysis for the bioactive potential of Streptomyces yeochonensis CN732, an acidophilic filamentous soil actinobacterium. BMC Genomics. 2020 Feb 3;21(1):118.
34. Jo HG, Adidjaja JJ, Kim DK, Park BS, Lee N, Cho BK, et al. Comparative genomic analysis of Streptomyces rapamycinicus NRRL 5491 and its mutant overproducing rapamycin. Sci Rep. 2022 Jun 18;12(1):10302.
35. Alam K, Hao J, Zhong L, Fan G, Ouyang Q, Islam MM, et al. Complete genome sequencing and in silico genome mining reveal the promising metabolic potential in Streptomyces strain CS-7. Front Microbiol. 2022 Oct 5;13:939919.
36. Um S, Guo H, Thiengmag S, Benndorf R, Murphy R, Rischer M, et al. Comparative Genomic and Metabolic Analysis of Streptomyces sp. RB110 Morphotypes Illuminates Genomic Rearrangements and Formation of a New 46-Membered Antimicrobial Macrolide. ACS Chem Biol. 2021 Aug 20;16(8):1482-1492.
37. Abdel-Glil MY, Thomas P, Linde J, Busch A, Wieler LH, Neubauer H, Seyboldt C. Comparative in silico genome analysis of Clostridium perfringens unravels stable phylogroups with different genome characteristics and pathogenic potential. Sci Rep. 2021 Mar 24;11(1):6756.
38. Kumar P, Chauhan A, Kumar M, Kuanr BK, Kundu A, Solanki R, Kapur MK. In vitro and in silico anticancer potential analysis of Streptomyces sp. extract against human lung cancer cell line, A549. 3 Biotech. 2021 Jun;11(6):254.
